Gene
KWMTBOMO09508
Pre Gene Modal
BGIBMGA014571
Annotation
PREDICTED:_serine_protease_gd-like_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.687 PlasmaMembrane Reliability : 1.25
Sequence
CDS
ATGGCTTTTATAAATATTATATCAATTAATAGTTGTATCCCTTCACTGCTGCTCATACTGCTGGTGTCAGCTCAAACGAAGACGCAGCCCGCGCTGGTGTCTCCGTGCCCTAGTGTCTTCGAATACGACACAAGCATACAGCATCCGGGTAGATGGTACGGCGTTATCAAGCTATCAAGCGACTACACTATCCACTCGTTGTGGTTGAACATACATCTGGACAACTATTCTGAGGCGCTTAAGAACTGTCTAGGGAGCGTGACGCAGGAGAACAAATACAAATTTATGATCAACAGAACGGATTTCGTAATCCACCCCGGCGAAGTGAAGAAAGTACAGATTTATGTCGAATATAACATTACGAGCTCTCCTCCTAAAATGATAGGTATAGTATTTAATGGTCGCAGAATATGCGGGCCGTCATTCGTTCTGTCCCTATTACATTCAGACTATCGTTATCACGGGACCGATATGCGGGGATGTCCTTATCGACCAAGCGCTCTGCCTTCCTCGGAGGAGGAAGCTTCCAGTAACCGAACATACAATATACAAAGCACTACCGACAGCGGCGTAGTTCCTAATCCAACAAACAAGACTGAAAACAAGGACGTTGACGATCTATTCGGCGGGCCAGTATCCTTTGTATTGCCGGATACAGTTAATAACTTTCACAACAAGAGCGAACGCAGCCGTTGTGGTAGAATATACGCAAAATCACCTTTGGTCATTGGAGAAGCGATCGAGACCATACATTGGCCTTGGCAGGTGTTAATATTAAGGTCGATAAGCGCTCATACATATCATATGACGGACTTCAAGTATCGGTGTAGCGGCACTCTGGTTTCGACGCGGCACGTGATCACGGCTGCTCATTGTCTCACGGCCCGGGGTCAGCTCATCGAAAGAGACCTCCAATTAATTGTTGCAGGGCGGCACCATTTGCACGTTCAATTAGTTAACGAAAAGCACATATTTGCTGAAATCAAAGATATAAAAATTCACCCGGAGTTCGACTACCAAACATTCAGAAACGACCTGAGCATTGTGGAGGCTGTGGATCGAATAAAATACTCTAGCTGGGTGCAGCCGGCGTGTCTCTGGCCGCAGACTCCGGTACCCGAGAAGCACCGCGCGAAAACTGGAGTGGTGGGGGGCTGGGGTTTCGATAAGTCAGGAGAACCAAATGAACTATTAACTACAATCGAAATGTCCGAGGTGGGCACGCAGTCCTGTCTTCAAGCTCACGATGCTTCGTTGGCGAACTACTTAACCGATAAGGTTTACTGCGCGAAATATGTCAATGGCACTAGTAACTGCCACAACGACAGCGGGGGCGGCATGATGGTCCGAGTGAAGAACAAATGGTACCTGCGCGGCGTGCTCTCGTTCTACTACTCGCCCGAGCTGCAGCACGCCTGTCAGAGTCACGCCCTATTCACGGACGTCGCCAAGCACTTGCCCTGGATCAAACAGCACGTCTACGACTTCTTGTGA
Protein
MAFINIISINSCIPSLLLILLVSAQTKTQPALVSPCPSVFEYDTSIQHPGRWYGVIKLSSDYTIHSLWLNIHLDNYSEALKNCLGSVTQENKYKFMINRTDFVIHPGEVKKVQIYVEYNITSSPPKMIGIVFNGRRICGPSFVLSLLHSDYRYHGTDMRGCPYRPSALPSSEEEASSNRTYNIQSTTDSGVVPNPTNKTENKDVDDLFGGPVSFVLPDTVNNFHNKSERSRCGRIYAKSPLVIGEAIETIHWPWQVLILRSISAHTYHMTDFKYRCSGTLVSTRHVITAAHCLTARGQLIERDLQLIVAGRHHLHVQLVNEKHIFAEIKDIKIHPEFDYQTFRNDLSIVEAVDRIKYSSWVQPACLWPQTPVPEKHRAKTGVVGGWGFDKSGEPNELLTTIEMSEVGTQSCLQAHDASLANYLTDKVYCAKYVNGTSNCHNDSGGGMMVRVKNKWYLRGVLSFYYSPELQHACQSHALFTDVAKHLPWIKQHVYDFL
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
EMBL
BABH01004022
BABH01004023
KZ150116
PZC73272.1
GEZM01087311
JAV58781.1
+ More
NEVH01014358 PNF27885.1 PNF27883.1 PNF27882.1 PNF27884.1 GEZM01037042 JAV82227.1 KK852507 KDR22462.1 LJIG01009271 KRT83413.1 KQ971357 EFA09211.1 KB632003 ERL87858.1 APGK01044224 APGK01044225 KB741021 ENN75080.1 GEZM01077642 GEZM01077641 JAV63217.1 LJIG01000649 KRT86291.1 KDR22461.1 KJ512078 AID60301.1 ENN75083.1
NEVH01014358 PNF27885.1 PNF27883.1 PNF27882.1 PNF27884.1 GEZM01037042 JAV82227.1 KK852507 KDR22462.1 LJIG01009271 KRT83413.1 KQ971357 EFA09211.1 KB632003 ERL87858.1 APGK01044224 APGK01044225 KB741021 ENN75080.1 GEZM01077642 GEZM01077641 JAV63217.1 LJIG01000649 KRT86291.1 KDR22461.1 KJ512078 AID60301.1 ENN75083.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF50494
SSF50494
CDD
ProteinModelPortal
PDB
4IW4
E-value=9.99655e-18,
Score=222
Ontologies
GO
Topology
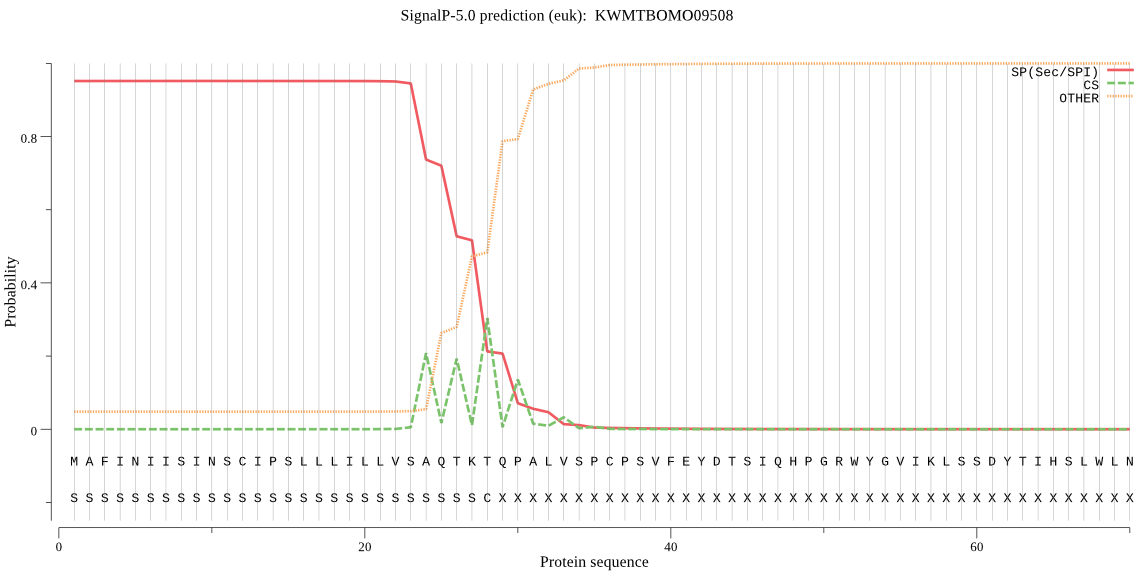
SignalP
Position: 1 - 28,
Likelihood: 0.952253
Length:
497
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
9.85603999999999
Exp number, first 60 AAs:
9.84699
Total prob of N-in:
0.45663
outside
1 - 497

Population Genetic Test Statistics
Pi
186.203286
Theta
162.219741
Tajima's D
0.339859
CLR
1.601268
CSRT
0.464076796160192
Interpretation
Uncertain
