Gene
KWMTBOMO09499
Pre Gene Modal
BGIBMGA013054
Annotation
ischemia/reperfusion_inducible_protein_[Bombyx_mori]
Full name
Threonylcarbamoyl-AMP synthase
Alternative Name
L-threonylcarbamoyladenylate synthase
Location in the cell
Mitochondrial Reliability : 3.098
Sequence
CDS
ATGTGCTCACAACTGGAGAGATTAAACTCATTGCGACTATTAACGAGTATGAAATTACTCAAACTTGGATTGTATCCTAGTTATATACGAAGAAATAAATGTCAATCTTTAATGATGAGAAACACAATGACATCTGTCATATCATGTGTAGATTTCACTGCAAGCTTGAGGGCTGCTGAATGCCTGGCTAAGGGCCAAGTGATTGCTGTACCGACAGACACAATCTATGGATTAGCTTGCAGTGCCAACTGTCCTCTGGCTATCAAAAAACTTTACTCTATAAAAGGAAGAGAGGCAGTGAAACCGGTGGCCATATGTGTCACTAGTGTAGCTGATTTGAGAAAATGGGGAGAAGCCTCTCACATAAGTGACAAGTTATTAGATTCGCTACTTCCTGGTCCAGTCACAGTTGTTCTACAGAAAACTAAACACCTAGACAATCCATTTTTGAATCCTTCCACTACAAAAATAGGTATTAGAATACCGAAGCATGATTTTATGAACACAGTTACGAAAATATTTGATATGCCAGTTGCATTGACAAGTGCCAATTTTAGTAATGAACCTTCAACATTGTCCATCAAAGAATTTGAACATCTGTATCCTCATTTAGCAGCCATATTTGATGGAGGGTTACTGAGTCAAGGTCTGGAACAGAACAGAACAGGCTCCACGGTTATAGATTTATCAAAGAATGGCCTTTATAAAATTATAAGAAAGGGTATATCATATGACAATGTTATAAAAGTTGTTGAACGTTATGGTTTGAAAAAAAATAATGAATCTGAAAATTCATTAGAGATAAATCAACAATTGCAGAAATAA
Protein
MCSQLERLNSLRLLTSMKLLKLGLYPSYIRRNKCQSLMMRNTMTSVISCVDFTASLRAAECLAKGQVIAVPTDTIYGLACSANCPLAIKKLYSIKGREAVKPVAICVTSVADLRKWGEASHISDKLLDSLLPGPVTVVLQKTKHLDNPFLNPSTTKIGIRIPKHDFMNTVTKIFDMPVALTSANFSNEPSTLSIKEFEHLYPHLAAIFDGGLLSQGLEQNRTGSTVIDLSKNGLYKIIRKGISYDNVIKVVERYGLKKNNESENSLEINQQLQK
Summary
Description
Required for the formation of a threonylcarbamoyl group on adenosine at position 37 (t(6)A37) in tRNAs that read codons beginning with adenine.
Catalytic Activity
ATP + hydrogencarbonate + L-threonine = diphosphate + H2O + L-threonylcarbamoyladenylate
Similarity
Belongs to the archaeal rpoM/eukaryotic RPA12/RPB9/RPC11 RNA polymerase family.
Belongs to the SUA5 family.
Belongs to the SUA5 family.
Uniprot
Q2F638
H9JU42
A0A2W1B6N6
A0A2A4JIU2
S4P972
A0A0L7LTN2
+ More
A0A194Q0I7 A0A0N1IDT4 A0A212FMX8 A0A023EMJ1 Q17L90 A0A2M4BWL8 A0A182FHI9 A0A0K8TSC5 W5JB92 A0A1Q3F4A0 A0A2M4AGY8 A0A182NI01 A0A2M4AJT7 A0A1J1I108 A0A2M4BWE8 B0X902 A0A1I8MQN2 T1DQE4 A0A0P9ACW7 A0A240PNZ4 A0A182XYR7 A0A2M4AEP0 A0A2M4AEQ1 A0A0R3NMZ9 A0A2M4ADT0 A0A0M4EJI0 A0A3B0IZU4 A0A1W4VHH9 B4MEA0 A0A2M3ZBG6 A0A2M3ZBE5 A0A1I8PQP7 B4MPJ1 B4KR44 B4J957 Q8SYJ9 Q7QJB7 A0A182V5U6 B4GGZ5 B4QF35 B3NJR9 A0A182LUH5 A0A182M6Q1 A0A1S3D309 A0A0A1WEB3 B4PA36 A0A0A1WMP6 A0A182WJC4 B4HQM8 A0A182KR71 A0A182UHI7 A0A1L8DHN9 A0A182PQJ6 A0A1L8DHW3 A0A443S6H7 A0A0K8WBE7 A0A182I364 E0VJY7 T2MAG2 U5D144 A0A1S3J0D6 A0A1Y9H9F2 A0A0D6R320 A0A1B6D014 A0A087SYA8 A0A1X7UH48 A0A2J7PHD8 E9H210 W5UDQ8 A0A287A202 K4AFB7 A0A452DV91 A0A1U7R2K0 W5QF38 A0A067QW52 T1HN26 A0A443RQ77 A0A1A6HTU9 J9K8U0 A4IG71 K4ADP5 A0A293M8X2 A0A0A0LUP4 A0A3B1JK95 F1R994 A0A1Y1JX87 A0A2Y9DCQ3 A0A0D9VQV4 A0A3B1IRF6 G1SNT6
A0A194Q0I7 A0A0N1IDT4 A0A212FMX8 A0A023EMJ1 Q17L90 A0A2M4BWL8 A0A182FHI9 A0A0K8TSC5 W5JB92 A0A1Q3F4A0 A0A2M4AGY8 A0A182NI01 A0A2M4AJT7 A0A1J1I108 A0A2M4BWE8 B0X902 A0A1I8MQN2 T1DQE4 A0A0P9ACW7 A0A240PNZ4 A0A182XYR7 A0A2M4AEP0 A0A2M4AEQ1 A0A0R3NMZ9 A0A2M4ADT0 A0A0M4EJI0 A0A3B0IZU4 A0A1W4VHH9 B4MEA0 A0A2M3ZBG6 A0A2M3ZBE5 A0A1I8PQP7 B4MPJ1 B4KR44 B4J957 Q8SYJ9 Q7QJB7 A0A182V5U6 B4GGZ5 B4QF35 B3NJR9 A0A182LUH5 A0A182M6Q1 A0A1S3D309 A0A0A1WEB3 B4PA36 A0A0A1WMP6 A0A182WJC4 B4HQM8 A0A182KR71 A0A182UHI7 A0A1L8DHN9 A0A182PQJ6 A0A1L8DHW3 A0A443S6H7 A0A0K8WBE7 A0A182I364 E0VJY7 T2MAG2 U5D144 A0A1S3J0D6 A0A1Y9H9F2 A0A0D6R320 A0A1B6D014 A0A087SYA8 A0A1X7UH48 A0A2J7PHD8 E9H210 W5UDQ8 A0A287A202 K4AFB7 A0A452DV91 A0A1U7R2K0 W5QF38 A0A067QW52 T1HN26 A0A443RQ77 A0A1A6HTU9 J9K8U0 A4IG71 K4ADP5 A0A293M8X2 A0A0A0LUP4 A0A3B1JK95 F1R994 A0A1Y1JX87 A0A2Y9DCQ3 A0A0D9VQV4 A0A3B1IRF6 G1SNT6
EC Number
2.7.7.87
Pubmed
19121390
28756777
23622113
26227816
26354079
22118469
+ More
24945155 17510324 26369729 20920257 23761445 25315136 17994087 25244985 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12364791 14747013 17210077 22936249 25830018 17550304 20966253 20566863 24065732 24357323 21292972 23127152 30723633 22580951 20809919 24845553 23594743 19881527 19495411 20565788 22047402 25329095 28004739 21993624
24945155 17510324 26369729 20920257 23761445 25315136 17994087 25244985 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12364791 14747013 17210077 22936249 25830018 17550304 20966253 20566863 24065732 24357323 21292972 23127152 30723633 22580951 20809919 24845553 23594743 19881527 19495411 20565788 22047402 25329095 28004739 21993624
EMBL
DQ311234
ABD36179.1
BABH01004030
KZ150346
PZC71278.1
NWSH01001411
+ More
PCG71330.1 GAIX01003859 JAA88701.1 JTDY01000111 KOB78797.1 KQ459582 KPI99066.1 KQ460651 KPJ13013.1 AGBW02007651 OWR55059.1 GAPW01003548 JAC10050.1 CH477217 EAT47485.1 GGFJ01008326 MBW57467.1 GDAI01000562 JAI17041.1 ADMH02001732 ETN61266.1 GFDL01012653 JAV22392.1 GGFK01006661 MBW39982.1 GGFK01007728 MBW41049.1 CVRI01000038 CRK94031.1 GGFJ01008222 MBW57363.1 DS232513 EDS42908.1 GAMD01001205 JAB00386.1 CH902619 KPU75915.1 GGFK01005923 MBW39244.1 GGFK01005929 MBW39250.1 CM000071 KRT02382.1 GGFK01005624 MBW38945.1 CP012524 ALC41417.1 OUUW01000001 SPP73825.1 CH940662 EDW58865.1 GGFM01005074 MBW25825.1 GGFM01005070 MBW25821.1 CH963849 EDW74030.1 CH933808 EDW08231.1 CH916367 EDW02432.1 AE013599 AY071501 AAF57437.2 AAL49123.1 AAAB01008807 EAA04653.3 CH479183 EDW35765.1 CM000362 CM002911 EDX07946.1 KMY95309.1 CH954179 EDV55338.1 AXCM01001984 AXCM01005915 GBXI01016963 JAC97328.1 CM000158 EDW91367.1 GBXI01014594 JAC99697.1 CH480816 EDW48731.1 GFDF01008118 JAV05966.1 GFDF01008117 JAV05967.1 NCKV01007048 RWS23137.1 GDHF01003917 JAI48397.1 APCN01000199 DS235235 EEB13693.1 HAAD01002857 CDG69089.1 KI392493 ERN15970.1 AXCN02000386 GCKF01027303 GCKF01027302 JAG98227.1 GEDC01018304 JAS18994.1 KK112514 KFM57847.1 NEVH01025141 PNF15742.1 GL732584 EFX74263.1 JT413219 AHH40161.1 AEMK02000045 DQIR01198614 HDB54091.1 AGNK02006093 LWLT01000004 AMGL01000860 AMGL01000861 KK852888 KDR14324.1 ACPB03015125 NCKU01000077 RWS17415.1 LZPO01010358 OBS81676.1 ABLF02033536 CU855952 FQ323133 BC134955 AAI34956.1 CM003536 RCV46048.1 GFWV01012505 MAA37234.1 CM002922 KGN64719.1 GEZM01102065 GEZM01102061 JAV51995.1
PCG71330.1 GAIX01003859 JAA88701.1 JTDY01000111 KOB78797.1 KQ459582 KPI99066.1 KQ460651 KPJ13013.1 AGBW02007651 OWR55059.1 GAPW01003548 JAC10050.1 CH477217 EAT47485.1 GGFJ01008326 MBW57467.1 GDAI01000562 JAI17041.1 ADMH02001732 ETN61266.1 GFDL01012653 JAV22392.1 GGFK01006661 MBW39982.1 GGFK01007728 MBW41049.1 CVRI01000038 CRK94031.1 GGFJ01008222 MBW57363.1 DS232513 EDS42908.1 GAMD01001205 JAB00386.1 CH902619 KPU75915.1 GGFK01005923 MBW39244.1 GGFK01005929 MBW39250.1 CM000071 KRT02382.1 GGFK01005624 MBW38945.1 CP012524 ALC41417.1 OUUW01000001 SPP73825.1 CH940662 EDW58865.1 GGFM01005074 MBW25825.1 GGFM01005070 MBW25821.1 CH963849 EDW74030.1 CH933808 EDW08231.1 CH916367 EDW02432.1 AE013599 AY071501 AAF57437.2 AAL49123.1 AAAB01008807 EAA04653.3 CH479183 EDW35765.1 CM000362 CM002911 EDX07946.1 KMY95309.1 CH954179 EDV55338.1 AXCM01001984 AXCM01005915 GBXI01016963 JAC97328.1 CM000158 EDW91367.1 GBXI01014594 JAC99697.1 CH480816 EDW48731.1 GFDF01008118 JAV05966.1 GFDF01008117 JAV05967.1 NCKV01007048 RWS23137.1 GDHF01003917 JAI48397.1 APCN01000199 DS235235 EEB13693.1 HAAD01002857 CDG69089.1 KI392493 ERN15970.1 AXCN02000386 GCKF01027303 GCKF01027302 JAG98227.1 GEDC01018304 JAS18994.1 KK112514 KFM57847.1 NEVH01025141 PNF15742.1 GL732584 EFX74263.1 JT413219 AHH40161.1 AEMK02000045 DQIR01198614 HDB54091.1 AGNK02006093 LWLT01000004 AMGL01000860 AMGL01000861 KK852888 KDR14324.1 ACPB03015125 NCKU01000077 RWS17415.1 LZPO01010358 OBS81676.1 ABLF02033536 CU855952 FQ323133 BC134955 AAI34956.1 CM003536 RCV46048.1 GFWV01012505 MAA37234.1 CM002922 KGN64719.1 GEZM01102065 GEZM01102061 JAV51995.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053268
UP000053240
UP000007151
+ More
UP000008820 UP000069272 UP000000673 UP000075884 UP000183832 UP000002320 UP000095301 UP000007801 UP000075880 UP000076408 UP000001819 UP000092553 UP000268350 UP000192221 UP000008792 UP000095300 UP000007798 UP000009192 UP000001070 UP000000803 UP000007062 UP000075903 UP000008744 UP000000304 UP000008711 UP000075883 UP000079169 UP000002282 UP000075920 UP000001292 UP000075882 UP000075902 UP000075885 UP000288716 UP000075840 UP000009046 UP000017836 UP000085678 UP000075886 UP000054359 UP000007879 UP000235965 UP000000305 UP000221080 UP000008227 UP000291000 UP000189706 UP000002356 UP000027135 UP000015103 UP000285301 UP000092124 UP000007819 UP000000437 UP000004995 UP000029981 UP000018467 UP000248480 UP000032180 UP000001811
UP000008820 UP000069272 UP000000673 UP000075884 UP000183832 UP000002320 UP000095301 UP000007801 UP000075880 UP000076408 UP000001819 UP000092553 UP000268350 UP000192221 UP000008792 UP000095300 UP000007798 UP000009192 UP000001070 UP000000803 UP000007062 UP000075903 UP000008744 UP000000304 UP000008711 UP000075883 UP000079169 UP000002282 UP000075920 UP000001292 UP000075882 UP000075902 UP000075885 UP000288716 UP000075840 UP000009046 UP000017836 UP000085678 UP000075886 UP000054359 UP000007879 UP000235965 UP000000305 UP000221080 UP000008227 UP000291000 UP000189706 UP000002356 UP000027135 UP000015103 UP000285301 UP000092124 UP000007819 UP000000437 UP000004995 UP000029981 UP000018467 UP000248480 UP000032180 UP000001811
Interpro
IPR017945
DHBP_synth_RibB-like_a/b_dom
+ More
IPR006070 YrdC-like_dom
IPR001529 DNA-dir_RNA_pol_M/15kDasu
IPR001222 Znf_TFIIS
IPR019761 DNA-dir_RNA_pol-M_15_CS
IPR034014 Zn_ribbon_RPC11_C
IPR036286 LexA/Signal_pep-like_sf
IPR000223 Pept_S26A_signal_pept_1
IPR015927 Peptidase_S24_S26A/B/C
IPR010923 t(6)A37_SUA5
IPR006070 YrdC-like_dom
IPR001529 DNA-dir_RNA_pol_M/15kDasu
IPR001222 Znf_TFIIS
IPR019761 DNA-dir_RNA_pol-M_15_CS
IPR034014 Zn_ribbon_RPC11_C
IPR036286 LexA/Signal_pep-like_sf
IPR000223 Pept_S26A_signal_pept_1
IPR015927 Peptidase_S24_S26A/B/C
IPR010923 t(6)A37_SUA5
ProteinModelPortal
Q2F638
H9JU42
A0A2W1B6N6
A0A2A4JIU2
S4P972
A0A0L7LTN2
+ More
A0A194Q0I7 A0A0N1IDT4 A0A212FMX8 A0A023EMJ1 Q17L90 A0A2M4BWL8 A0A182FHI9 A0A0K8TSC5 W5JB92 A0A1Q3F4A0 A0A2M4AGY8 A0A182NI01 A0A2M4AJT7 A0A1J1I108 A0A2M4BWE8 B0X902 A0A1I8MQN2 T1DQE4 A0A0P9ACW7 A0A240PNZ4 A0A182XYR7 A0A2M4AEP0 A0A2M4AEQ1 A0A0R3NMZ9 A0A2M4ADT0 A0A0M4EJI0 A0A3B0IZU4 A0A1W4VHH9 B4MEA0 A0A2M3ZBG6 A0A2M3ZBE5 A0A1I8PQP7 B4MPJ1 B4KR44 B4J957 Q8SYJ9 Q7QJB7 A0A182V5U6 B4GGZ5 B4QF35 B3NJR9 A0A182LUH5 A0A182M6Q1 A0A1S3D309 A0A0A1WEB3 B4PA36 A0A0A1WMP6 A0A182WJC4 B4HQM8 A0A182KR71 A0A182UHI7 A0A1L8DHN9 A0A182PQJ6 A0A1L8DHW3 A0A443S6H7 A0A0K8WBE7 A0A182I364 E0VJY7 T2MAG2 U5D144 A0A1S3J0D6 A0A1Y9H9F2 A0A0D6R320 A0A1B6D014 A0A087SYA8 A0A1X7UH48 A0A2J7PHD8 E9H210 W5UDQ8 A0A287A202 K4AFB7 A0A452DV91 A0A1U7R2K0 W5QF38 A0A067QW52 T1HN26 A0A443RQ77 A0A1A6HTU9 J9K8U0 A4IG71 K4ADP5 A0A293M8X2 A0A0A0LUP4 A0A3B1JK95 F1R994 A0A1Y1JX87 A0A2Y9DCQ3 A0A0D9VQV4 A0A3B1IRF6 G1SNT6
A0A194Q0I7 A0A0N1IDT4 A0A212FMX8 A0A023EMJ1 Q17L90 A0A2M4BWL8 A0A182FHI9 A0A0K8TSC5 W5JB92 A0A1Q3F4A0 A0A2M4AGY8 A0A182NI01 A0A2M4AJT7 A0A1J1I108 A0A2M4BWE8 B0X902 A0A1I8MQN2 T1DQE4 A0A0P9ACW7 A0A240PNZ4 A0A182XYR7 A0A2M4AEP0 A0A2M4AEQ1 A0A0R3NMZ9 A0A2M4ADT0 A0A0M4EJI0 A0A3B0IZU4 A0A1W4VHH9 B4MEA0 A0A2M3ZBG6 A0A2M3ZBE5 A0A1I8PQP7 B4MPJ1 B4KR44 B4J957 Q8SYJ9 Q7QJB7 A0A182V5U6 B4GGZ5 B4QF35 B3NJR9 A0A182LUH5 A0A182M6Q1 A0A1S3D309 A0A0A1WEB3 B4PA36 A0A0A1WMP6 A0A182WJC4 B4HQM8 A0A182KR71 A0A182UHI7 A0A1L8DHN9 A0A182PQJ6 A0A1L8DHW3 A0A443S6H7 A0A0K8WBE7 A0A182I364 E0VJY7 T2MAG2 U5D144 A0A1S3J0D6 A0A1Y9H9F2 A0A0D6R320 A0A1B6D014 A0A087SYA8 A0A1X7UH48 A0A2J7PHD8 E9H210 W5UDQ8 A0A287A202 K4AFB7 A0A452DV91 A0A1U7R2K0 W5QF38 A0A067QW52 T1HN26 A0A443RQ77 A0A1A6HTU9 J9K8U0 A4IG71 K4ADP5 A0A293M8X2 A0A0A0LUP4 A0A3B1JK95 F1R994 A0A1Y1JX87 A0A2Y9DCQ3 A0A0D9VQV4 A0A3B1IRF6 G1SNT6
PDB
1JCU
E-value=1.32817e-19,
Score=235
Ontologies
GO
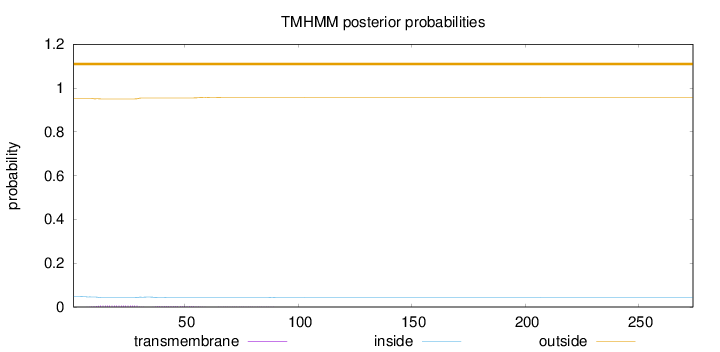
Topology
Subcellular location
Cytoplasm
Length:
274
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.20541
Exp number, first 60 AAs:
0.18838
Total prob of N-in:
0.04716
outside
1 - 274
Population Genetic Test Statistics
Pi
39.361708
Theta
23.90013
Tajima's D
-0.606868
CLR
0
CSRT
0.220488975551222
Interpretation
Uncertain
