Gene
KWMTBOMO09498
Pre Gene Modal
BGIBMGA012810
Annotation
hypothetical_protein_KGM_15428_[Danaus_plexippus]
Full name
Mitochondrial inner membrane protease subunit
+ More
Mitochondrial inner membrane protease subunit 2
Mitochondrial inner membrane protease subunit 2
Alternative Name
IMP2-like protein
Location in the cell
Extracellular Reliability : 2.271
Sequence
CDS
ATGTGGCTTAAAAGTGTATGCAAGTCTCTTGTCTTCGGCTTACCAATCGGTGTAACCATTCTAGACACGGTCGGTTATGTGGCTAGAGTTGAAGGAATTTCTATGCAACCCGTGCTGAATCCAGAATCGATGAATACGGACTATGTGTTTTTATCTCGATGGGCCGTCAGAGATTATCACGTGAAACGAGGAGATGTCATTTCACTGATGTCCCCCAAAGATCCTAACCAAAAGATTATCAAACGAGTGGTAGCTTTACAAGGTGACGTTGTAAGCACATTGGGCTACAAGAACCAGTATGTGAAAATTCCAGAAGGACATTGTTGGGTAGAAGGTGATCACACAGGTCACACCCTCGACAGCAACACCTTTGGGCCTGTTTCTTTAGGTTTAGTTAATGCAAGAGCGGTGTGCATAGTTTGGCCCCCAAGTAGATGGCAGAGTCTTCAAGCTAAATTGCCAGAGAACAGACAGCCTGTGAGCACTGCTATTTAA
Protein
MWLKSVCKSLVFGLPIGVTILDTVGYVARVEGISMQPVLNPESMNTDYVFLSRWAVRDYHVKRGDVISLMSPKDPNQKIIKRVVALQGDVVSTLGYKNQYVKIPEGHCWVEGDHTGHTLDSNTFGPVSLGLVNARAVCIVWPPSRWQSLQAKLPENRQPVSTAI
Summary
Description
Catalyzes the removal of transit peptides required for the targeting of proteins from the mitochondrial matrix, across the inner membrane, into the inter-membrane space.
Subunit
Heterodimer of 2 subunits, IMMPL1 and IMMPL2.
Similarity
Belongs to the peptidase S26 family.
Belongs to the peptidase S26 family. IMP2 subfamily.
Belongs to the peptidase S26 family. IMP2 subfamily.
Keywords
Complete proteome
Hydrolase
Membrane
Mitochondrion
Mitochondrion inner membrane
Protease
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain Mitochondrial inner membrane protease subunit 2
Uniprot
H9JTE8
A0A212FMZ3
A0A0L7LTJ6
A0A2H1WY76
A0A2W1BEL8
A0A2A4JIV1
+ More
I4DLG4 A0A0N0PC74 A0A2M3ZBJ2 A0A2M3ZAZ7 A0A0L0CRL4 A0A084WN42 W5JEL3 A0A2M4C0P4 A0A2M4C0A2 A0A182JQ88 A0A182I365 A0A1Q3FKG3 A0A182X3V7 Q7PT24 A0A182WJC3 D6WFF2 A0A182PQJ5 A0A182NI00 A0A034WY78 B0X901 A0A0K8WBY1 A0A1Y9HB30 A0A182MF66 A0A1I8P0G3 A0A1I8N279 T1DFT7 A0A0T6ATM8 Q17L88 A0A1W7R7V3 A0A023EIT0 A0A1A9WWF6 B4MEA1 A0A1Y1KWW9 A0A2J7R985 A0A1A9USA7 A0A1A9ZAU7 D3TPQ8 A0A1A9XIW5 A0A1B0C050 V5I7H1 B4PA37 B4KR45 B3MIU0 B3NJR8 B4HQM7 Q4QQ12 A0A0K8TLA9 B4MPJ0 B4J956 A0A067QTG8 B4QF34 A0A1B6HAG0 A0A1W4VGW9 Q17L89 N6T7C2 A0A3B0JKQ7 B4GGZ6 Q28ZP5 U4U0Z2 A0A1L8DTV8 A0A1L8DTZ8 A0A336KE86 V9LE80 A0A232F250 K7JK06 A0A023FA07 A0A224XME0 A0A3Q1BJB1 A0A3P8TKV0 T1JHL3 A0A3Q1H8K8 A0A250XVM3 R4FK57 A0A3Q3EPD1 A0A1S3MKA1 F1R3I2 Q6AZD4 A0A3B4T286 A0A1S2ZA20 A0A0S7M451 G3NZJ6 E2BY07 A0A2R7VY59 K9IH67 F7DWS9 A0A0P6JDQ8 A0A3Q7PFQ0 A0A2K5Q8G8 W5UK72 U3AWW4 K9K261 A0A1A7XXJ6 G3NZK0 A0A2K6SCW0
I4DLG4 A0A0N0PC74 A0A2M3ZBJ2 A0A2M3ZAZ7 A0A0L0CRL4 A0A084WN42 W5JEL3 A0A2M4C0P4 A0A2M4C0A2 A0A182JQ88 A0A182I365 A0A1Q3FKG3 A0A182X3V7 Q7PT24 A0A182WJC3 D6WFF2 A0A182PQJ5 A0A182NI00 A0A034WY78 B0X901 A0A0K8WBY1 A0A1Y9HB30 A0A182MF66 A0A1I8P0G3 A0A1I8N279 T1DFT7 A0A0T6ATM8 Q17L88 A0A1W7R7V3 A0A023EIT0 A0A1A9WWF6 B4MEA1 A0A1Y1KWW9 A0A2J7R985 A0A1A9USA7 A0A1A9ZAU7 D3TPQ8 A0A1A9XIW5 A0A1B0C050 V5I7H1 B4PA37 B4KR45 B3MIU0 B3NJR8 B4HQM7 Q4QQ12 A0A0K8TLA9 B4MPJ0 B4J956 A0A067QTG8 B4QF34 A0A1B6HAG0 A0A1W4VGW9 Q17L89 N6T7C2 A0A3B0JKQ7 B4GGZ6 Q28ZP5 U4U0Z2 A0A1L8DTV8 A0A1L8DTZ8 A0A336KE86 V9LE80 A0A232F250 K7JK06 A0A023FA07 A0A224XME0 A0A3Q1BJB1 A0A3P8TKV0 T1JHL3 A0A3Q1H8K8 A0A250XVM3 R4FK57 A0A3Q3EPD1 A0A1S3MKA1 F1R3I2 Q6AZD4 A0A3B4T286 A0A1S2ZA20 A0A0S7M451 G3NZJ6 E2BY07 A0A2R7VY59 K9IH67 F7DWS9 A0A0P6JDQ8 A0A3Q7PFQ0 A0A2K5Q8G8 W5UK72 U3AWW4 K9K261 A0A1A7XXJ6 G3NZK0 A0A2K6SCW0
EC Number
3.4.21.-
Pubmed
19121390
22118469
26227816
28756777
22651552
26354079
+ More
26108605 24438588 20920257 23761445 12364791 14747013 17210077 18362917 19820115 25348373 25315136 17510324 24945155 17994087 28004739 20353571 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26369729 24845553 22936249 23537049 15632085 24402279 28648823 20075255 25474469 28087693 23594743 20798317 19892987 23127152 25243066
26108605 24438588 20920257 23761445 12364791 14747013 17210077 18362917 19820115 25348373 25315136 17510324 24945155 17994087 28004739 20353571 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26369729 24845553 22936249 23537049 15632085 24402279 28648823 20075255 25474469 28087693 23594743 20798317 19892987 23127152 25243066
EMBL
BABH01004030
AGBW02007651
OWR55060.1
JTDY01000111
KOB78803.1
ODYU01011974
+ More
SOQ58023.1 KZ150346 PZC71280.1 NWSH01001411 PCG71329.1 PCG71332.1 AK402132 KQ459582 BAM18754.1 KPI99065.1 KQ460651 KPJ13014.1 GGFM01005122 MBW25873.1 GGFM01004963 MBW25714.1 JRES01000105 KNC34079.1 ATLV01024562 KE525352 KFB51636.1 ADMH02001732 ETN61264.1 GGFJ01009600 MBW58741.1 GGFJ01009604 MBW58745.1 APCN01000199 GFDL01006997 JAV28048.1 AAAB01008807 EAA04666.4 KQ971319 EFA00254.1 GAKP01000236 GAKP01000234 JAC58718.1 DS232513 EDS42907.1 GDHF01024721 GDHF01016685 GDHF01003821 GDHF01003722 JAI27593.1 JAI35629.1 JAI48493.1 JAI48592.1 AXCN02000386 AXCM01005915 GAMD01003036 JAA98554.1 LJIG01022848 KRT78406.1 CH477217 EAT47487.1 GEHC01000403 JAV47242.1 GAPW01004733 JAC08865.1 CH940662 EDW58866.1 GEZM01071281 JAV65913.1 NEVH01006600 PNF37400.1 EZ423410 ADD19686.1 JXJN01023463 GALX01006445 JAB62021.1 CM000158 EDW91368.1 CH933808 EDW08232.1 CH902619 EDV36000.1 CH954179 EDV55337.1 CH480816 EDW48730.1 AE013599 BT023604 AAF57438.1 AAY85004.1 GDAI01002898 JAI14705.1 CH963849 EDW74029.1 CH916367 EDW02431.1 KK852954 KDR13303.1 CM000362 CM002911 EDX07945.1 KMY95308.1 GECU01036042 JAS71664.1 EAT47486.1 APGK01041186 KB740991 ENN76099.1 OUUW01000001 SPP73826.1 CH479183 EDW35766.1 CM000071 EAL25568.1 KB631348 KB631467 ERL84251.1 ERL84312.1 GFDF01004206 JAV09878.1 GFDF01004224 JAV09860.1 UFQS01000316 UFQT01000289 UFQT01000316 SSX02767.1 SSX22844.1 JW878144 AFP10661.1 NNAY01001170 OXU24896.1 GBBI01000954 GEMB01002187 JAC17758.1 JAS00993.1 GFTR01002769 JAW13657.1 JH431612 GFFV01004535 JAV35410.1 ACPB03006700 GAHY01002018 JAA75492.1 CR354561 CR384055 CR759824 BC078193 GBYX01119923 JAO96478.1 GL451363 EFN79441.1 KK854163 PTY12392.1 GABZ01008223 JAA45302.1 GEBF01001103 JAO02530.1 JT416626 AHH42100.1 GAMT01007108 GAMS01011261 GAMQ01007138 GAMP01002857 JAB04753.1 JAB11875.1 JAB34713.1 JAB49898.1 JL616023 AEH58230.1 HADW01009630 HADX01000579 SBP22811.1
SOQ58023.1 KZ150346 PZC71280.1 NWSH01001411 PCG71329.1 PCG71332.1 AK402132 KQ459582 BAM18754.1 KPI99065.1 KQ460651 KPJ13014.1 GGFM01005122 MBW25873.1 GGFM01004963 MBW25714.1 JRES01000105 KNC34079.1 ATLV01024562 KE525352 KFB51636.1 ADMH02001732 ETN61264.1 GGFJ01009600 MBW58741.1 GGFJ01009604 MBW58745.1 APCN01000199 GFDL01006997 JAV28048.1 AAAB01008807 EAA04666.4 KQ971319 EFA00254.1 GAKP01000236 GAKP01000234 JAC58718.1 DS232513 EDS42907.1 GDHF01024721 GDHF01016685 GDHF01003821 GDHF01003722 JAI27593.1 JAI35629.1 JAI48493.1 JAI48592.1 AXCN02000386 AXCM01005915 GAMD01003036 JAA98554.1 LJIG01022848 KRT78406.1 CH477217 EAT47487.1 GEHC01000403 JAV47242.1 GAPW01004733 JAC08865.1 CH940662 EDW58866.1 GEZM01071281 JAV65913.1 NEVH01006600 PNF37400.1 EZ423410 ADD19686.1 JXJN01023463 GALX01006445 JAB62021.1 CM000158 EDW91368.1 CH933808 EDW08232.1 CH902619 EDV36000.1 CH954179 EDV55337.1 CH480816 EDW48730.1 AE013599 BT023604 AAF57438.1 AAY85004.1 GDAI01002898 JAI14705.1 CH963849 EDW74029.1 CH916367 EDW02431.1 KK852954 KDR13303.1 CM000362 CM002911 EDX07945.1 KMY95308.1 GECU01036042 JAS71664.1 EAT47486.1 APGK01041186 KB740991 ENN76099.1 OUUW01000001 SPP73826.1 CH479183 EDW35766.1 CM000071 EAL25568.1 KB631348 KB631467 ERL84251.1 ERL84312.1 GFDF01004206 JAV09878.1 GFDF01004224 JAV09860.1 UFQS01000316 UFQT01000289 UFQT01000316 SSX02767.1 SSX22844.1 JW878144 AFP10661.1 NNAY01001170 OXU24896.1 GBBI01000954 GEMB01002187 JAC17758.1 JAS00993.1 GFTR01002769 JAW13657.1 JH431612 GFFV01004535 JAV35410.1 ACPB03006700 GAHY01002018 JAA75492.1 CR354561 CR384055 CR759824 BC078193 GBYX01119923 JAO96478.1 GL451363 EFN79441.1 KK854163 PTY12392.1 GABZ01008223 JAA45302.1 GEBF01001103 JAO02530.1 JT416626 AHH42100.1 GAMT01007108 GAMS01011261 GAMQ01007138 GAMP01002857 JAB04753.1 JAB11875.1 JAB34713.1 JAB49898.1 JL616023 AEH58230.1 HADW01009630 HADX01000579 SBP22811.1
Proteomes
UP000005204
UP000007151
UP000037510
UP000218220
UP000053268
UP000053240
+ More
UP000037069 UP000030765 UP000000673 UP000075881 UP000075840 UP000076407 UP000007062 UP000075920 UP000007266 UP000075885 UP000075884 UP000002320 UP000075886 UP000075883 UP000095300 UP000095301 UP000008820 UP000091820 UP000008792 UP000235965 UP000078200 UP000092445 UP000092443 UP000092460 UP000002282 UP000009192 UP000007801 UP000008711 UP000001292 UP000000803 UP000007798 UP000001070 UP000027135 UP000000304 UP000192221 UP000019118 UP000268350 UP000008744 UP000001819 UP000030742 UP000215335 UP000002358 UP000257160 UP000265080 UP000257200 UP000015103 UP000261660 UP000087266 UP000000437 UP000261420 UP000079721 UP000007635 UP000008237 UP000002281 UP000286641 UP000233040 UP000221080 UP000008225 UP000233220
UP000037069 UP000030765 UP000000673 UP000075881 UP000075840 UP000076407 UP000007062 UP000075920 UP000007266 UP000075885 UP000075884 UP000002320 UP000075886 UP000075883 UP000095300 UP000095301 UP000008820 UP000091820 UP000008792 UP000235965 UP000078200 UP000092445 UP000092443 UP000092460 UP000002282 UP000009192 UP000007801 UP000008711 UP000001292 UP000000803 UP000007798 UP000001070 UP000027135 UP000000304 UP000192221 UP000019118 UP000268350 UP000008744 UP000001819 UP000030742 UP000215335 UP000002358 UP000257160 UP000265080 UP000257200 UP000015103 UP000261660 UP000087266 UP000000437 UP000261420 UP000079721 UP000007635 UP000008237 UP000002281 UP000286641 UP000233040 UP000221080 UP000008225 UP000233220
Interpro
SUPFAM
SSF51306
SSF51306
ProteinModelPortal
H9JTE8
A0A212FMZ3
A0A0L7LTJ6
A0A2H1WY76
A0A2W1BEL8
A0A2A4JIV1
+ More
I4DLG4 A0A0N0PC74 A0A2M3ZBJ2 A0A2M3ZAZ7 A0A0L0CRL4 A0A084WN42 W5JEL3 A0A2M4C0P4 A0A2M4C0A2 A0A182JQ88 A0A182I365 A0A1Q3FKG3 A0A182X3V7 Q7PT24 A0A182WJC3 D6WFF2 A0A182PQJ5 A0A182NI00 A0A034WY78 B0X901 A0A0K8WBY1 A0A1Y9HB30 A0A182MF66 A0A1I8P0G3 A0A1I8N279 T1DFT7 A0A0T6ATM8 Q17L88 A0A1W7R7V3 A0A023EIT0 A0A1A9WWF6 B4MEA1 A0A1Y1KWW9 A0A2J7R985 A0A1A9USA7 A0A1A9ZAU7 D3TPQ8 A0A1A9XIW5 A0A1B0C050 V5I7H1 B4PA37 B4KR45 B3MIU0 B3NJR8 B4HQM7 Q4QQ12 A0A0K8TLA9 B4MPJ0 B4J956 A0A067QTG8 B4QF34 A0A1B6HAG0 A0A1W4VGW9 Q17L89 N6T7C2 A0A3B0JKQ7 B4GGZ6 Q28ZP5 U4U0Z2 A0A1L8DTV8 A0A1L8DTZ8 A0A336KE86 V9LE80 A0A232F250 K7JK06 A0A023FA07 A0A224XME0 A0A3Q1BJB1 A0A3P8TKV0 T1JHL3 A0A3Q1H8K8 A0A250XVM3 R4FK57 A0A3Q3EPD1 A0A1S3MKA1 F1R3I2 Q6AZD4 A0A3B4T286 A0A1S2ZA20 A0A0S7M451 G3NZJ6 E2BY07 A0A2R7VY59 K9IH67 F7DWS9 A0A0P6JDQ8 A0A3Q7PFQ0 A0A2K5Q8G8 W5UK72 U3AWW4 K9K261 A0A1A7XXJ6 G3NZK0 A0A2K6SCW0
I4DLG4 A0A0N0PC74 A0A2M3ZBJ2 A0A2M3ZAZ7 A0A0L0CRL4 A0A084WN42 W5JEL3 A0A2M4C0P4 A0A2M4C0A2 A0A182JQ88 A0A182I365 A0A1Q3FKG3 A0A182X3V7 Q7PT24 A0A182WJC3 D6WFF2 A0A182PQJ5 A0A182NI00 A0A034WY78 B0X901 A0A0K8WBY1 A0A1Y9HB30 A0A182MF66 A0A1I8P0G3 A0A1I8N279 T1DFT7 A0A0T6ATM8 Q17L88 A0A1W7R7V3 A0A023EIT0 A0A1A9WWF6 B4MEA1 A0A1Y1KWW9 A0A2J7R985 A0A1A9USA7 A0A1A9ZAU7 D3TPQ8 A0A1A9XIW5 A0A1B0C050 V5I7H1 B4PA37 B4KR45 B3MIU0 B3NJR8 B4HQM7 Q4QQ12 A0A0K8TLA9 B4MPJ0 B4J956 A0A067QTG8 B4QF34 A0A1B6HAG0 A0A1W4VGW9 Q17L89 N6T7C2 A0A3B0JKQ7 B4GGZ6 Q28ZP5 U4U0Z2 A0A1L8DTV8 A0A1L8DTZ8 A0A336KE86 V9LE80 A0A232F250 K7JK06 A0A023FA07 A0A224XME0 A0A3Q1BJB1 A0A3P8TKV0 T1JHL3 A0A3Q1H8K8 A0A250XVM3 R4FK57 A0A3Q3EPD1 A0A1S3MKA1 F1R3I2 Q6AZD4 A0A3B4T286 A0A1S2ZA20 A0A0S7M451 G3NZJ6 E2BY07 A0A2R7VY59 K9IH67 F7DWS9 A0A0P6JDQ8 A0A3Q7PFQ0 A0A2K5Q8G8 W5UK72 U3AWW4 K9K261 A0A1A7XXJ6 G3NZK0 A0A2K6SCW0
PDB
6B88
E-value=0.0221545,
Score=83
Ontologies
GO
PANTHER
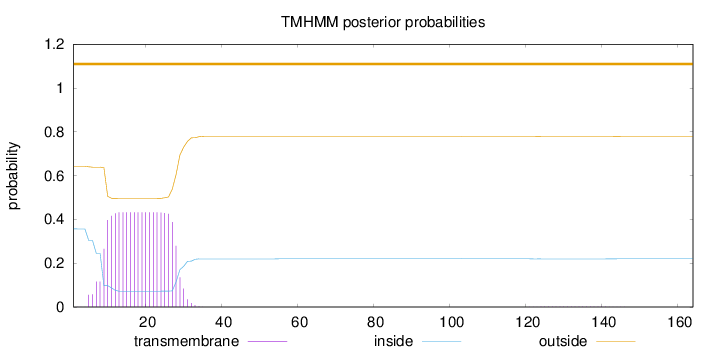
Topology
Subcellular location
Mitochondrion inner membrane
Length:
164
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
8.88527000000001
Exp number, first 60 AAs:
8.85054000000001
Total prob of N-in:
0.35725
outside
1 - 164
Population Genetic Test Statistics
Pi
87.604192
Theta
165.600264
Tajima's D
-1.481308
CLR
763.534486
CSRT
0.0627468626568672
Interpretation
Uncertain
