Pre Gene Modal
BGIBMGA014503
Annotation
PREDICTED:_RRP12-like_protein_[Amyelois_transitella]
Full name
RRP12-like protein
Location in the cell
Nuclear Reliability : 2.347
Sequence
CDS
ATGGGTAAATTTAGATCAAAACTTAAAGGCAAAACGAAGGGGAAACGATGGCAGAAAGGACAGTCATCAAATTCGAACCCGAAAATTCAAAAATACAGGGAGATGGCAAAATCCCGGTTTTTTCAGGAAAACTTAGGCAGCACTGGCTTAACACAGCAAGCGGTGTCCAAGCATGAAGCTATGATGACATATGGACATAAAACTAACAAAACAAAATCAGATGCTGAAAATGAACTGACCATAGTAAAAGAGCTTGAAAATATGTCGGTCAAGACCGGAGACGAGGGATCAGAGTCTGAATACTCTGGTACATTAAGATCAGGAACATTTAAGACATTTCAAACATTTGCATCTGATTGGAGTCAATGTTCAAATATCAGCTTTCATAAATTGCTAAATCGTTTTGATTCAAACAATGCACTGCACAAGGAAATGCTAGCAGTGCTGGCGGCTGTGACTGAAGTTATCAAAGAACAAGGAGGTAAAGAGACTACAACGGAATACTTTGCTGCTCTTATGGAAACTCTAAAGGCATCCGAACTGGAAGACAATTTAGTAGCCACAATAACTTTACTATCAATGGGCATCAAAGCAGTACCACAAGCAGTACTGCGTAAACAATTTTCGGACTCCTCAACAATATTTATGGAATTACTCGAGGAGCATGCTCAGTCTGAAAATGGAGCATTGCTGAGGAGCACAATTGGATGTCTGTCGGTGCTGCTGAGAGCTCAGGAGTATGCTGTCTGGGGTGATTCATCAACTATGAGAGTGTTTGAATCCGTTCTGGCCTTTGCACTGCACTCTAAACCCAAGGTTAGAAAAGCGGCACAACATGCTGTGGTATCTATACTACGCGGAAGTAGCTTCATGATACCATCAGAGGATAAAAAAATTAAGGTACCGAAGTTGCATCCAGCTTCGAACCACGTGGCCGAGTACTGTCTTTCCCAGATCAAAGCAGAGGCGCTGCTCTCCGGTCATCGTCCTGTGTTGCACACACTGACCATGCTACGAGATGTGTTACCGGTGTTCAGCAAGGACAGTATAAAGACTATATGCGAGGCTATCTTATCTCTAATGACCCACAACAATGTCTACATCAAGACATGCTGTCTCCATACACTGCACTCTCTGTTTGCTGCACCACTGGACACTTGTAACTTGACGAGCGGGCTGGCTGTGCAGTTGACCAGAGCCTTAATGGCTTCCAGGCCCCCGGCCAACGATACAGGGCAGGTCTTGGCCTGGGCAGCTGTTTTGCAGCAGGGATATGGGTGTTTAGCTAATCTAGATTTGAATCTGTGCGTGCCAAATTTGCCGTCATTTGTAAACATATGCGTATCGGAGCTATGGCTGTCCGATGTTGCCAGCATTAATTCAGCTTCTACGAATGCACTTAAGGCCATCCTGCACGACTGCGTGAAGCCGGCCATTGAATCCGATGAAATATTCGCGAAGCACAAATCTCACGTAGAGGGTGTTTTCAAAGTGATCGGAGCGGGCCTGGACAATCCCTTCAATCAAGCCATCAGACACGTCATATTGACTATCGCCGTCTGTTTCGAGATAGCCAACGAAAAGATATCGAGCACCCTCGAGCCTCTGCTGAGGAAACTGAACGATCGTCGGGAAAGCCACAACTTCCACAACGACAAGGAAGTCGAACACGCAACCGGATGCGCCATACGGAGCCTGGGTCCGGAGTTTGTTCTCAAAACTATTCCGTTACGAGACAACCCTGATTCGATAAACCTGGAACGGAGCTGGCTGCTGCCGGTTTTGAAAGAAAAAATAACGCATTCGAATTTAAAATATTTCGCAACGGAAATTTTGGAGATGGCAACATTCTGCAGGAAGAAGTCTAGGGAGCTGTCACAGGCCGGCGACGTGCCGGGCTCGCACACGTACGAACTGCTCTGCAATCAATTCTGGGCTTTGCTGCCCAGTTTCTGCAATAGCCCGAAGGATATCAAGGACACCTTCAAGTTCATGGCCAGAGTGCTAGGTAACGTTCTCAAAGACAATCCGGAGTTCAGACTGTCGGTGATGCAGGCTCTGCGGAAACTCATCGCTTGCTCGTTAGAAAACGACGAGGACAAGCTAGAGATGAGCAGATACGCGAAAAACTACCTCCCGATCCTGCTGAACGTATACATGTCTCCTGCCAAGGGCAGCAGCGCGGAAGGACAGAGATTAGCCGCTCTGGAGACCATACAGGTATATCTAAGTATATCGGACGAGAACCTCAACCGAGAACTGTTCGACAACGCCGTGAAGCAATTGGAAGCAGCACGAGACGACCACTTCCAGAGAGAGTCGATATTGGACGTGATAAGAGTGTTGGTACTGTACCAAGAGAGCGATCGAATCGCGAAACTGTTCGACGACTGGCTGTACCCGCTGTGCGAGACCGTGCTGGAGGACCCGAGGACCTTCAAGAAGGCCAAGAAGAAAGCCGCAGCCATGGAGGAAGACTCCGCTGATAAGAAACCCGAACTCCGCTACAAGGACAAGGCGCGTCTCCTCGAGATGGAGCACAAGAAAGCCTACAGAATACTGGAAGAGATATTCAAGAGCGAGAAGGCTCCATGCAAGGAGTTCCTTCGCTCGAACCACAAGAAGATAAAGAAGTTGCTGATGACGTCACTGAACAAAGTCGCCGACAGCAGCAAGGCCGCCAGATTGAGGTGCCTAGAGTGCTTGATCAACACGACGCCGACCCTGAACGCGGAGAGCAAGCTGCTGAGGAGTGCTATAGCCGAGTCCGTTGTCTGCACGAGAGATATTAATTCGAAATGTCGCCAGTGCGCCTTCAATCTGATCACGTGCGTCGGGAACGTGCTGAAGAAGCAAGATGGAGGTACGCCTTTTTTATATATGGCTTAG
Protein
MGKFRSKLKGKTKGKRWQKGQSSNSNPKIQKYREMAKSRFFQENLGSTGLTQQAVSKHEAMMTYGHKTNKTKSDAENELTIVKELENMSVKTGDEGSESEYSGTLRSGTFKTFQTFASDWSQCSNISFHKLLNRFDSNNALHKEMLAVLAAVTEVIKEQGGKETTTEYFAALMETLKASELEDNLVATITLLSMGIKAVPQAVLRKQFSDSSTIFMELLEEHAQSENGALLRSTIGCLSVLLRAQEYAVWGDSSTMRVFESVLAFALHSKPKVRKAAQHAVVSILRGSSFMIPSEDKKIKVPKLHPASNHVAEYCLSQIKAEALLSGHRPVLHTLTMLRDVLPVFSKDSIKTICEAILSLMTHNNVYIKTCCLHTLHSLFAAPLDTCNLTSGLAVQLTRALMASRPPANDTGQVLAWAAVLQQGYGCLANLDLNLCVPNLPSFVNICVSELWLSDVASINSASTNALKAILHDCVKPAIESDEIFAKHKSHVEGVFKVIGAGLDNPFNQAIRHVILTIAVCFEIANEKISSTLEPLLRKLNDRRESHNFHNDKEVEHATGCAIRSLGPEFVLKTIPLRDNPDSINLERSWLLPVLKEKITHSNLKYFATEILEMATFCRKKSRELSQAGDVPGSHTYELLCNQFWALLPSFCNSPKDIKDTFKFMARVLGNVLKDNPEFRLSVMQALRKLIACSLENDEDKLEMSRYAKNYLPILLNVYMSPAKGSSAEGQRLAALETIQVYLSISDENLNRELFDNAVKQLEAARDDHFQRESILDVIRVLVLYQESDRIAKLFDDWLYPLCETVLEDPRTFKKAKKKAAAMEEDSADKKPELRYKDKARLLEMEHKKAYRILEEIFKSEKAPCKEFLRSNHKKIKKLLMTSLNKVADSSKAARLRCLECLINTTPTLNAESKLLRSAIAESVVCTRDINSKCRQCAFNLITCVGNVLKKQDGGTPFLYMA
Summary
Similarity
Belongs to the RRP12 family.
Keywords
Complete proteome
Nucleus
Phosphoprotein
Reference proteome
Feature
chain RRP12-like protein
Uniprot
H9JY83
A0A2H1W140
A0A437BFC6
A0A2W1B8U5
A0A212FMT2
A0A0N0PC57
+ More
A0A194Q165 K7IVE6 A0A232EV45 N6URZ0 A0A2J7PYA8 A0A0C9QA54 D6WZI8 A0A067R5V1 N6UP81 A0A1Q3G0S0 W4VRP4 Q16NI7 A0A1Q3G0P6 A0A0L7RJQ9 A0A088ADE3 A0A084WP17 A0A0D3QC34 A0A0D3QBW7 T1PKZ6 A0A0D3QBX7 A0A0D3QCH0 A0A0D3QCH2 A0A0D3QBW8 A0A0D3QC38 A0A0D3QBX4 A0A0D3QCM9 A0A0D3QBX0 A0A0D3QCP0 A0A182U002 A0A182HU17 A0A0D3QBW3 A0A182G4E5 Q7QD52 A0A0D3QBX1 A0A182WRR1 A0A1I8N8S8 A0A0D3QCN5 A0A0D3QBY0 A0A0D3QC25 A0A0D3QCI3 A0A0D3QC30 A0A0D3QCP5 A0A0D3QBW6 A0A0L0C2M7 A0A1I8NRM7 A0A182P6M9 W5JGT3 A0A2M3ZFM3 A0A2M4B9I0 A0A2M4CZ58 A0A2M4B9G9 A0A182FPG6 A0A2M4B9E6 A0A182XWC5 A0A182ISQ2 A0A182K671 A0A182LG11 A0A1B0CT14 A0A1W4VH78 B0XAB9 E2BNF3 A0A0K8U887 B4Q2E0 A0A2M4A4H5 A0A2M4A4V3 A0A151IMR5 E2AJV9 A0A182VYK1 A0A0A1X1A8 A0A2A3EDN3 X2JEW2 A0A182S835 Q9VYA7 C9QPD2 W8AUA0 A0A2M4A4K2 A0A1B6HBS5 A0A034VB23 A0A182NDX0 C5WLN0 A0A182M725 A0A0J7NVB3 A0A195AT88 A0A158NPQ6 A0A182RNB5 A0A195EL12 B3N0A0 B3NWL5 A0A182QJL6 A0A0M4ENF4 A0A1A9WXZ1 Q29FM7
A0A194Q165 K7IVE6 A0A232EV45 N6URZ0 A0A2J7PYA8 A0A0C9QA54 D6WZI8 A0A067R5V1 N6UP81 A0A1Q3G0S0 W4VRP4 Q16NI7 A0A1Q3G0P6 A0A0L7RJQ9 A0A088ADE3 A0A084WP17 A0A0D3QC34 A0A0D3QBW7 T1PKZ6 A0A0D3QBX7 A0A0D3QCH0 A0A0D3QCH2 A0A0D3QBW8 A0A0D3QC38 A0A0D3QBX4 A0A0D3QCM9 A0A0D3QBX0 A0A0D3QCP0 A0A182U002 A0A182HU17 A0A0D3QBW3 A0A182G4E5 Q7QD52 A0A0D3QBX1 A0A182WRR1 A0A1I8N8S8 A0A0D3QCN5 A0A0D3QBY0 A0A0D3QC25 A0A0D3QCI3 A0A0D3QC30 A0A0D3QCP5 A0A0D3QBW6 A0A0L0C2M7 A0A1I8NRM7 A0A182P6M9 W5JGT3 A0A2M3ZFM3 A0A2M4B9I0 A0A2M4CZ58 A0A2M4B9G9 A0A182FPG6 A0A2M4B9E6 A0A182XWC5 A0A182ISQ2 A0A182K671 A0A182LG11 A0A1B0CT14 A0A1W4VH78 B0XAB9 E2BNF3 A0A0K8U887 B4Q2E0 A0A2M4A4H5 A0A2M4A4V3 A0A151IMR5 E2AJV9 A0A182VYK1 A0A0A1X1A8 A0A2A3EDN3 X2JEW2 A0A182S835 Q9VYA7 C9QPD2 W8AUA0 A0A2M4A4K2 A0A1B6HBS5 A0A034VB23 A0A182NDX0 C5WLN0 A0A182M725 A0A0J7NVB3 A0A195AT88 A0A158NPQ6 A0A182RNB5 A0A195EL12 B3N0A0 B3NWL5 A0A182QJL6 A0A0M4ENF4 A0A1A9WXZ1 Q29FM7
Pubmed
19121390
28756777
22118469
26354079
20075255
28648823
+ More
23537049 18362917 19820115 24845553 17510324 24438588 25552603 26483478 12364791 14747013 17210077 25315136 26108605 20920257 23761445 25244985 20966253 20798317 17994087 17550304 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 17372656 18327897 24495485 25348373 21347285 15632085
23537049 18362917 19820115 24845553 17510324 24438588 25552603 26483478 12364791 14747013 17210077 25315136 26108605 20920257 23761445 25244985 20966253 20798317 17994087 17550304 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 17372656 18327897 24495485 25348373 21347285 15632085
EMBL
BABH01042523
BABH01042524
BABH01042525
BABH01042526
ODYU01005702
SOQ46815.1
+ More
RSAL01000071 RVE49136.1 KZ150346 PZC71281.1 AGBW02007651 OWR55062.1 KQ460651 KPJ13017.1 KQ459582 KPI99063.1 AAZX01004302 NNAY01002059 OXU22207.1 APGK01001802 KB734468 ENN83556.1 NEVH01020850 PNF21318.1 GBYB01000109 JAG69876.1 KQ971372 EFA09698.2 KK852722 KDR17709.1 ENN83555.1 GFDL01001663 JAV33382.1 GANO01000610 JAB59261.1 CH477823 EAT35907.1 GFDL01001666 JAV33379.1 KQ414581 KOC71033.1 ATLV01024815 KE525361 KFB51961.1 KP274278 AJC98362.1 KP274264 AJC98348.1 KA648790 AFP63419.1 KP274279 AJC98363.1 KP274266 AJC98350.1 KP274271 AJC98355.1 KP274277 AJC98361.1 KP274283 AJC98367.1 KP274274 AJC98358.1 KP274265 AJC98349.1 KP274282 AJC98366.1 KP274275 AJC98359.1 APCN01000599 KP274267 KP274276 AJC98351.1 AJC98360.1 JXUM01143584 KQ569626 KXJ68574.1 AAAB01008859 EAA07536.4 KP274269 AJC98353.1 KP274270 AJC98354.1 KP274284 AJC98368.1 KP274268 AJC98352.1 KP274281 AJC98365.1 KP274273 AJC98357.1 KP274280 AJC98364.1 KP274272 AJC98356.1 JRES01000984 KNC26487.1 ADMH02001267 ETN63296.1 GGFM01006616 MBW27367.1 GGFJ01000510 MBW49651.1 GGFL01006435 MBW70613.1 GGFJ01000511 MBW49652.1 GGFJ01000512 MBW49653.1 AJWK01026873 AJWK01026874 AJWK01026875 DS232577 EDS43616.1 GL449414 EFN82732.1 GDHF01029548 JAI22766.1 CM000162 EDX01601.1 GGFK01002338 MBW35659.1 GGFK01002439 MBW35760.1 KQ977026 KYN06248.1 GL440100 EFN66275.1 GBXI01009874 GBXI01004258 JAD04418.1 JAD10034.1 KZ288271 PBC29810.1 AE014298 AHN59688.1 AY051753 BT100014 ACX54922.1 GAMC01018272 JAB88283.1 GGFK01002339 MBW35660.1 GECU01035585 JAS72121.1 GAKP01020221 GAKP01020219 JAC38733.1 BT088845 ACS54297.1 AXCM01005292 LBMM01001428 KMQ96335.1 KQ976745 KYM75438.1 ADTU01022709 KQ978730 KYN28945.1 CH902640 EDV38304.1 CH954180 EDV47177.2 AXCN02001589 CP012528 ALC49496.1 CH379066 EAL31373.3
RSAL01000071 RVE49136.1 KZ150346 PZC71281.1 AGBW02007651 OWR55062.1 KQ460651 KPJ13017.1 KQ459582 KPI99063.1 AAZX01004302 NNAY01002059 OXU22207.1 APGK01001802 KB734468 ENN83556.1 NEVH01020850 PNF21318.1 GBYB01000109 JAG69876.1 KQ971372 EFA09698.2 KK852722 KDR17709.1 ENN83555.1 GFDL01001663 JAV33382.1 GANO01000610 JAB59261.1 CH477823 EAT35907.1 GFDL01001666 JAV33379.1 KQ414581 KOC71033.1 ATLV01024815 KE525361 KFB51961.1 KP274278 AJC98362.1 KP274264 AJC98348.1 KA648790 AFP63419.1 KP274279 AJC98363.1 KP274266 AJC98350.1 KP274271 AJC98355.1 KP274277 AJC98361.1 KP274283 AJC98367.1 KP274274 AJC98358.1 KP274265 AJC98349.1 KP274282 AJC98366.1 KP274275 AJC98359.1 APCN01000599 KP274267 KP274276 AJC98351.1 AJC98360.1 JXUM01143584 KQ569626 KXJ68574.1 AAAB01008859 EAA07536.4 KP274269 AJC98353.1 KP274270 AJC98354.1 KP274284 AJC98368.1 KP274268 AJC98352.1 KP274281 AJC98365.1 KP274273 AJC98357.1 KP274280 AJC98364.1 KP274272 AJC98356.1 JRES01000984 KNC26487.1 ADMH02001267 ETN63296.1 GGFM01006616 MBW27367.1 GGFJ01000510 MBW49651.1 GGFL01006435 MBW70613.1 GGFJ01000511 MBW49652.1 GGFJ01000512 MBW49653.1 AJWK01026873 AJWK01026874 AJWK01026875 DS232577 EDS43616.1 GL449414 EFN82732.1 GDHF01029548 JAI22766.1 CM000162 EDX01601.1 GGFK01002338 MBW35659.1 GGFK01002439 MBW35760.1 KQ977026 KYN06248.1 GL440100 EFN66275.1 GBXI01009874 GBXI01004258 JAD04418.1 JAD10034.1 KZ288271 PBC29810.1 AE014298 AHN59688.1 AY051753 BT100014 ACX54922.1 GAMC01018272 JAB88283.1 GGFK01002339 MBW35660.1 GECU01035585 JAS72121.1 GAKP01020221 GAKP01020219 JAC38733.1 BT088845 ACS54297.1 AXCM01005292 LBMM01001428 KMQ96335.1 KQ976745 KYM75438.1 ADTU01022709 KQ978730 KYN28945.1 CH902640 EDV38304.1 CH954180 EDV47177.2 AXCN02001589 CP012528 ALC49496.1 CH379066 EAL31373.3
Proteomes
UP000005204
UP000283053
UP000007151
UP000053240
UP000053268
UP000002358
+ More
UP000215335 UP000019118 UP000235965 UP000007266 UP000027135 UP000008820 UP000053825 UP000005203 UP000030765 UP000075902 UP000075840 UP000069940 UP000249989 UP000007062 UP000076407 UP000095301 UP000075903 UP000037069 UP000095300 UP000075885 UP000000673 UP000069272 UP000076408 UP000075880 UP000075881 UP000075882 UP000092461 UP000192221 UP000002320 UP000008237 UP000002282 UP000078542 UP000000311 UP000075920 UP000242457 UP000000803 UP000075901 UP000075884 UP000075883 UP000036403 UP000078540 UP000005205 UP000075900 UP000078492 UP000007801 UP000008711 UP000075886 UP000092553 UP000091820 UP000001819
UP000215335 UP000019118 UP000235965 UP000007266 UP000027135 UP000008820 UP000053825 UP000005203 UP000030765 UP000075902 UP000075840 UP000069940 UP000249989 UP000007062 UP000076407 UP000095301 UP000075903 UP000037069 UP000095300 UP000075885 UP000000673 UP000069272 UP000076408 UP000075880 UP000075881 UP000075882 UP000092461 UP000192221 UP000002320 UP000008237 UP000002282 UP000078542 UP000000311 UP000075920 UP000242457 UP000000803 UP000075901 UP000075884 UP000075883 UP000036403 UP000078540 UP000005205 UP000075900 UP000078492 UP000007801 UP000008711 UP000075886 UP000092553 UP000091820 UP000001819
Pfam
PF08161 NUC173
SUPFAM
SSF48371
SSF48371
Gene 3D
ProteinModelPortal
H9JY83
A0A2H1W140
A0A437BFC6
A0A2W1B8U5
A0A212FMT2
A0A0N0PC57
+ More
A0A194Q165 K7IVE6 A0A232EV45 N6URZ0 A0A2J7PYA8 A0A0C9QA54 D6WZI8 A0A067R5V1 N6UP81 A0A1Q3G0S0 W4VRP4 Q16NI7 A0A1Q3G0P6 A0A0L7RJQ9 A0A088ADE3 A0A084WP17 A0A0D3QC34 A0A0D3QBW7 T1PKZ6 A0A0D3QBX7 A0A0D3QCH0 A0A0D3QCH2 A0A0D3QBW8 A0A0D3QC38 A0A0D3QBX4 A0A0D3QCM9 A0A0D3QBX0 A0A0D3QCP0 A0A182U002 A0A182HU17 A0A0D3QBW3 A0A182G4E5 Q7QD52 A0A0D3QBX1 A0A182WRR1 A0A1I8N8S8 A0A0D3QCN5 A0A0D3QBY0 A0A0D3QC25 A0A0D3QCI3 A0A0D3QC30 A0A0D3QCP5 A0A0D3QBW6 A0A0L0C2M7 A0A1I8NRM7 A0A182P6M9 W5JGT3 A0A2M3ZFM3 A0A2M4B9I0 A0A2M4CZ58 A0A2M4B9G9 A0A182FPG6 A0A2M4B9E6 A0A182XWC5 A0A182ISQ2 A0A182K671 A0A182LG11 A0A1B0CT14 A0A1W4VH78 B0XAB9 E2BNF3 A0A0K8U887 B4Q2E0 A0A2M4A4H5 A0A2M4A4V3 A0A151IMR5 E2AJV9 A0A182VYK1 A0A0A1X1A8 A0A2A3EDN3 X2JEW2 A0A182S835 Q9VYA7 C9QPD2 W8AUA0 A0A2M4A4K2 A0A1B6HBS5 A0A034VB23 A0A182NDX0 C5WLN0 A0A182M725 A0A0J7NVB3 A0A195AT88 A0A158NPQ6 A0A182RNB5 A0A195EL12 B3N0A0 B3NWL5 A0A182QJL6 A0A0M4ENF4 A0A1A9WXZ1 Q29FM7
A0A194Q165 K7IVE6 A0A232EV45 N6URZ0 A0A2J7PYA8 A0A0C9QA54 D6WZI8 A0A067R5V1 N6UP81 A0A1Q3G0S0 W4VRP4 Q16NI7 A0A1Q3G0P6 A0A0L7RJQ9 A0A088ADE3 A0A084WP17 A0A0D3QC34 A0A0D3QBW7 T1PKZ6 A0A0D3QBX7 A0A0D3QCH0 A0A0D3QCH2 A0A0D3QBW8 A0A0D3QC38 A0A0D3QBX4 A0A0D3QCM9 A0A0D3QBX0 A0A0D3QCP0 A0A182U002 A0A182HU17 A0A0D3QBW3 A0A182G4E5 Q7QD52 A0A0D3QBX1 A0A182WRR1 A0A1I8N8S8 A0A0D3QCN5 A0A0D3QBY0 A0A0D3QC25 A0A0D3QCI3 A0A0D3QC30 A0A0D3QCP5 A0A0D3QBW6 A0A0L0C2M7 A0A1I8NRM7 A0A182P6M9 W5JGT3 A0A2M3ZFM3 A0A2M4B9I0 A0A2M4CZ58 A0A2M4B9G9 A0A182FPG6 A0A2M4B9E6 A0A182XWC5 A0A182ISQ2 A0A182K671 A0A182LG11 A0A1B0CT14 A0A1W4VH78 B0XAB9 E2BNF3 A0A0K8U887 B4Q2E0 A0A2M4A4H5 A0A2M4A4V3 A0A151IMR5 E2AJV9 A0A182VYK1 A0A0A1X1A8 A0A2A3EDN3 X2JEW2 A0A182S835 Q9VYA7 C9QPD2 W8AUA0 A0A2M4A4K2 A0A1B6HBS5 A0A034VB23 A0A182NDX0 C5WLN0 A0A182M725 A0A0J7NVB3 A0A195AT88 A0A158NPQ6 A0A182RNB5 A0A195EL12 B3N0A0 B3NWL5 A0A182QJL6 A0A0M4ENF4 A0A1A9WXZ1 Q29FM7
Ontologies
GO
Topology
Subcellular location
Nucleus
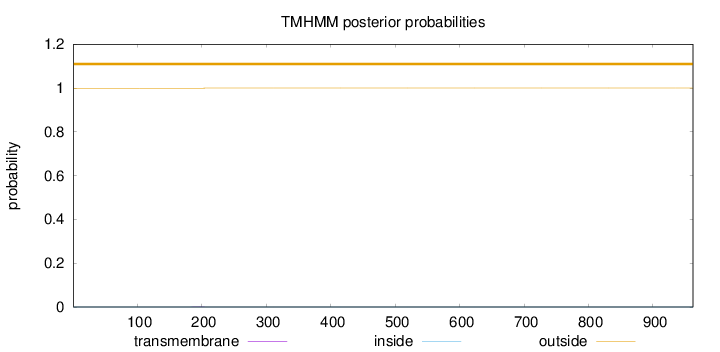
Length:
962
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00854999999999998
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.00044
outside
1 - 962
Population Genetic Test Statistics
Pi
296.861049
Theta
150.840743
Tajima's D
2.623072
CLR
0.30126
CSRT
0.95945202739863
Interpretation
Uncertain
