Gene
KWMTBOMO09492 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012807
Annotation
PREDICTED:_carboxypeptidase_D_isoform_X3_[Bombyx_mori]
Full name
Carboxypeptidase D
Alternative Name
Metallocarboxypeptidase D
Protein silver
Protein silver
Location in the cell
Cytoplasmic Reliability : 1.25 Nuclear Reliability : 1.135
Sequence
CDS
ATGTATCGCTGGATTCGCATGTTGATACTGCTGTTTTCTCTAACAATCGCCAAAGCACAAGTTTCAGCAGTCACAAATGAGGACGAATCCTTTCTAAAGACATCAAATTATACGAAATACGACCAACTTGGTATTCTATTTGACAAACTCGAAAGCACCTATCCGGATCTGGCGAAGTTATATTCGATTGGGAATTCCGTCGAAGGTAGGAAGTTGCTGGTACTACAAATAACGCAGGACGTGCAGAATGAACACCCGGAACGTCCAGCGTTTAAATACGTGGCAAATATGCACGGCGATGAATCGATTGGAAGACAAATAGTGATATATTTGGCTCAGTACTTGTTATTGAACTATGGTAAAAATGAAAGGGTCACGAAGCTGGTGAACAGCACAGATATACACTTGATGCCATCTCTGAACCCGGATGGGTTTGAGGCTAGTGAGGAAGGAAACTGCGAATCGTTAAGAGGCTACGTCGGTCGTAGTAATGCAAACGGCGTGGACTTGAACCGCGATTTTCCCGACCAGTTCGACAAGAATCATGCGAACGATGACGAATATTTATATAGCGGACGGCAACCGGAGACCGTGGCCCTCATCCGGTGGGTCCTGAACAAACAGTTTGTGCTGTCGGCTAACCTTCATGGAGGGGCTATTGTGGCGAGCTATCCATACGACGACGTAGGGCACGGAAAGAACAACGGGAAGGATTGTTGCGAAGAGAGCAGGTCGCCCGACGATGCCGTGTTCAGACACCTGGCCAATTCGTACGCGTCTAGGAACGCGGACATGAGGGCCGGGGACCGATGTAAGCCGGAGAACTTCACTAATGGCATCACCAACGGCGCTAACTGGTATTTCGTGCAGGGTGGAATGCAAGACTTTAATTACATCCACTCGAACTGCTTCGAGGTGACCTTCGAGCTCTCCTGCTGCAAGTACCCGCGCGCCGAGGAACTGCCCTCGTATTGGCACATGAACAAGGAGGCGATGGTCGCGTTCGTCGAGCAGGCTCACATCGGCATCAGCGGCGTGGTGCTGGACGAGGACGGCGCACCTGTGAAGGACGCGCATATCTTAGTGGACGGCATCAAACACCCGGTCAGCACCACGCAGCACGGCCGCTACTGGCGACTCCTCACTCCCGGACAGTACAATGTCACTGTCAAAGCTTCAGGGTTCGTTTCCCCACCGCCCACAACGGTGACCGTCCCAGAAAACCAGACCGCCGCCGTCCCGCTCAATCTGACCGTCCACCGTCGGCCGCGTTCGATAGTCGACGAGGATTTTGTCCATCACAATTACGAGAAGATGGAGTCTTACCTCAAGGACCTCAACGCGATGTACTCCAATATCACGAAGCTGACCAGCATCGGCAAATCGGTGCAAGGAAGAGAGTTATACGTACTGGAGGTTACCAGAGATCCAGGCGTGCATATTCCCGGGAAACCGGAATTCAAGTACGTAGCCAATATGCATGGAAACGAAGCCGTCGGACGCGAACTTCTGCTGTTGTTAGCGAAGTACTTGTGTCAGCAGTACGTCGCCGGCGATGAACGAATACAGAACATGCTGAACAACACGAGGATCCACTTGCTGCCGAGCATGAACCCTGACGGCTACGAGACTGCCAGACCTGGTGATTTCGATGGTTTGAAGGGTCGCACTAATGCACACGATGTTGACTTGAACAGGAATTTCCCCGATCAGTTTGGAAGTACCGAGGAAAATGAAGTCCAAGAGCCGGAGACGTCGGCGGTGATGAAGTGGTCGCAGTCGATCCCGTTCGTGCTGTCGGCCAACCTGCACGGCGGCGCGCTGGTCGCCAACTACCCCTACGACGACAACCCTAGCATGCGCAGCGGGAAGGAGTTCCTGTCTCCCGACAACCCTGTCTTCGTCCATCTCGCTCATGTTTACTCGGACGCCCACCTCAAGATGCACTTAGGGAAGCCCTGCAAGAACTACTCCAGGGAAATGTTCCCGGACGGCACCACGAACGGAGCTAAGTGGTACGTGCTCGCGGGCGGCATGCAAGATTGGAACTACCTGCACACAGACGACATGGAGCTCACGCTAGAGCTGGGCTGCTACAAGTTCCCTCCTGCCGCGGACCTTCCCACCTACTGGCAGGACAACAGGGAAGCTCTCTTGCAGTACATAGAACAGGTCCACACCGGAGTCCACGGCTTCGTCTATAGCCACATAGGTCACGGCTTGTCCAACGCCACGATAACTGTGTCCGGGATCAAGCACGCGGTCCACACGGCCCGGGCCGGCGACTACTGGCGGCTGCTGAGGCCGGGCGTGTACAACGTCACCGCCAGCAGACAGGGTTATGAGAGTGTGACGGAGGAGGTGACGGTCCCTCCCGCCGGCGCGGTCAGCCTCAACTTCACGCTGATGGCGGACGACTCGCAGCACTGGTCCTCCGCCTACGACTTCCGTATATTGGATAACGTTTTGAACACGATGTACCACACCCCTCTGCAAATGTACGCGGCCCTATCTGAATTGGAGAACAAGTATCCGAGCATAGCGGAGTTCCGGGCCGGGGACAGCCTGGTCACCTCCAAGTTCCATCAGCTCAAGATGACTTCGAATGTCGGCTCTCCAGAGGAGACCAAATTCCACATTGCGATAATGAGCAGCCTGTACGCGACCCAGCCTCTGGGCCGGGAGATGCTTCTGAACTTTGCACGTCACATCGCCACCGCCTACACCATCGGCGAACCGAAACACCAACGCCTACTGAACAACACCGTCCTCCACTTCATACCGAACATCGACCCGCTCAACTCGAAGATAATCAAGCAATACGACGGCACCGACAAGTGCGACCTCACCGCCCTCGAGGAAGAGTTCGGCGATAGCCTGTACAACTACCTGACGAAGACCAACATGAACCCACTCTCGAATTACACCAGCGAAACGGCCTTCATCGAAATGCTCAAATCGGAGAAGTACGACTTGATTCTAGAACTCTCTTCCGGCACAGAAGACGTGTCTTACCCTGAAGCTTCGAAGGAACTGTACGAGAAATACGCCCGGACGTACCAGGACCACAGGAACCCAAGCGATAAGTACGAGTGTCGGCCGTTGGACAGCAGTGTCCACGGTCAGCTCGTGGACTTGCTGTGCGAGCGGCTCGACGTGCCCGTGGTGTCCGTGGGGTTGAGCTGCTGCAGGATGCCCGTCGCCGACCAGATCGGCTGGGTCTGGAGAGACAACCTGGGCGGCATCATGAAGTTCGTCGAGCAAGCCAATACCGGCGTCGTGGGATACGTAAAGAACGAGATGGGAAAGCCGGTCCGCGACGCCACCATAGCCATCACCAGCTTCTCGAAGCAGTACCGGGTGACGAGCAACATGGCGCACTACCACGTGATGCTGCCCGCGGGCGTCTACAAGGTCATCGTGCGCGCTCACAACTACAAGGATCGGATCCTAACTTGGAACGTCCTGGACTCCAAACTGAAGCAAATCGACATAATACTGCAGAGGACGAATGCGGAGAGGATATCTGGTGGTCAATTCGAGGACATCCAAGTTCCAGAGGATCCTAATGTCGTTTATACCATGGGGCTGGTCCTGTCCCGCAGCAGCGAGCCGCTGGGCGGGGCCGAGCTGGCGGTGCGCGGGGCGCGCGCGGGCGGCGTGCTGGCGCGCAACACCAGCGACGCGCACGGCCGCTTCGTGCTCGCGCTGCCCGCCGGGTACAAGGGGAAGCAGGTCGCGATATCCGCCGCCTCCGACGGATACGTCACCAAGCAGATGGACGTCTTGATCAACAGCGTGGACAACTTAACGCCCAATTTGATATTTAAGCTGGACAGCGACGACAATGTTCTAGGGATGCCACGTCTCGTTTTCGTCATGTTGTCTGGCGTGGTGGGCGTGTCGCTGGTGGTGCTGGGGGCGTGGTGCTTCAGCTGCCGCGAGCGCGCCCGCCACTCGCGCCGCGACTACCTGTTCACGCAGCTGCCCGGGGACGACAAGCGCCCGCTCTGCGAGCCTGATTTGATCCGCAAGCCGTTCTACGACGAGGAGGACATCCCGTTGACGGAGACCGACTCAGAGGAAGATGTCGTGCTGCTGCAGACCGGACGAGAGCAATGA
Protein
MYRWIRMLILLFSLTIAKAQVSAVTNEDESFLKTSNYTKYDQLGILFDKLESTYPDLAKLYSIGNSVEGRKLLVLQITQDVQNEHPERPAFKYVANMHGDESIGRQIVIYLAQYLLLNYGKNERVTKLVNSTDIHLMPSLNPDGFEASEEGNCESLRGYVGRSNANGVDLNRDFPDQFDKNHANDDEYLYSGRQPETVALIRWVLNKQFVLSANLHGGAIVASYPYDDVGHGKNNGKDCCEESRSPDDAVFRHLANSYASRNADMRAGDRCKPENFTNGITNGANWYFVQGGMQDFNYIHSNCFEVTFELSCCKYPRAEELPSYWHMNKEAMVAFVEQAHIGISGVVLDEDGAPVKDAHILVDGIKHPVSTTQHGRYWRLLTPGQYNVTVKASGFVSPPPTTVTVPENQTAAVPLNLTVHRRPRSIVDEDFVHHNYEKMESYLKDLNAMYSNITKLTSIGKSVQGRELYVLEVTRDPGVHIPGKPEFKYVANMHGNEAVGRELLLLLAKYLCQQYVAGDERIQNMLNNTRIHLLPSMNPDGYETARPGDFDGLKGRTNAHDVDLNRNFPDQFGSTEENEVQEPETSAVMKWSQSIPFVLSANLHGGALVANYPYDDNPSMRSGKEFLSPDNPVFVHLAHVYSDAHLKMHLGKPCKNYSREMFPDGTTNGAKWYVLAGGMQDWNYLHTDDMELTLELGCYKFPPAADLPTYWQDNREALLQYIEQVHTGVHGFVYSHIGHGLSNATITVSGIKHAVHTARAGDYWRLLRPGVYNVTASRQGYESVTEEVTVPPAGAVSLNFTLMADDSQHWSSAYDFRILDNVLNTMYHTPLQMYAALSELENKYPSIAEFRAGDSLVTSKFHQLKMTSNVGSPEETKFHIAIMSSLYATQPLGREMLLNFARHIATAYTIGEPKHQRLLNNTVLHFIPNIDPLNSKIIKQYDGTDKCDLTALEEEFGDSLYNYLTKTNMNPLSNYTSETAFIEMLKSEKYDLILELSSGTEDVSYPEASKELYEKYARTYQDHRNPSDKYECRPLDSSVHGQLVDLLCERLDVPVVSVGLSCCRMPVADQIGWVWRDNLGGIMKFVEQANTGVVGYVKNEMGKPVRDATIAITSFSKQYRVTSNMAHYHVMLPAGVYKVIVRAHNYKDRILTWNVLDSKLKQIDIILQRTNAERISGGQFEDIQVPEDPNVVYTMGLVLSRSSEPLGGAELAVRGARAGGVLARNTSDAHGRFVLALPAGYKGKQVAISAASDGYVTKQMDVLINSVDNLTPNLIFKLDSDDNVLGMPRLVFVMLSGVVGVSLVVLGAWCFSCRERARHSRRDYLFTQLPGDDKRPLCEPDLIRKPFYDEEDIPLTETDSEEDVVLLQTGREQ
Summary
Description
Required for the proper melanization and sclerotization of the cuticle.
Catalytic Activity
Releases C-terminal Arg and Lys from polypeptides.
Cofactor
Zn(2+)
Similarity
Belongs to the peptidase M14 family.
Keywords
Alternative splicing
Carboxypeptidase
Complete proteome
Cuticle
Glycoprotein
Hydrolase
Membrane
Metal-binding
Metalloprotease
Phosphoprotein
Protease
Reference proteome
Repeat
Signal
Transmembrane
Transmembrane helix
Zinc
Feature
chain Carboxypeptidase D
splice variant In isoform 3, isoform 4 and isoform 6.
splice variant In isoform 3, isoform 4 and isoform 6.
Uniprot
H9JTE5
A0A3S2M1L9
A0A212FMU7
A0A1Y1NB52
A0A1L8DR43
A0A1L8DQU3
+ More
A0A1W6EVY0 A0A182P717 A0A2J7RM97 F4X480 W4VRN2 A0A067RRP6 A0A151I9W8 A0A3L8E7V4 A0A348G601 A0A1V1FVK8 A0A182QQ91 A0A158N9I3 E2AE72 A0A151I452 A0A182VX80 A0A026WDN1 A0A0L7R5M1 A0A182YKF6 A0A2A3EQH4 A0A087ZRN5 A0A0J7KJS1 A0A151JV41 A0A151XGD6 A0A0M3QZA1 A0A182HT57 A0A182NA58 A0A182UMV6 Q7QC23 A0A182XI70 A0A195E1K9 A0A182RI04 E2BNR3 A0A0Q9WMF1 J9HJF8 A0A1W4UVK5 A0A0L0BTV6 A0A1I8Q4W2 A0A182J566 B4PX05 B4MEQ9 B4R7E8 A0A182F709 A0A1Q3G2F0 B3P973 A0A1Q3G2L4 W5JS53 A0A154PFK1 A0A1Q3G2Y6 A0A1Q3G2Y9 A0A1Q3G243 A0A2M4B9D9 P42787-5 A0A1Q3G254 X2J9Z1 A0A2M4B9F1 A0A2M4A298 A0A0P8ZHH2 A0A2M4A2G0 B4L2T3 A0A2M4B9B6 P42787 A0A1W4V999 A0A3B0KTY1 A0A0A1XAK1 A0A1A9UUA2 B4JNA3 B5DMP1 A0A1J1IBI2 A0A0R1EF72 B4GY07 A0A1B6LU96 A0A0K8VVD6 V9IFV8 A0A0Q5SRQ2 A0A1B6BYT4 P42787-4 P42787-3 B4N2H9 B3MYX9 A0A0Q9W7V5 A0A1I8MWK5 A0A2M3YZY6 A0A2M3YZV8 A0A310SMJ0 A0A0R3P055 A0A1B6GCF9 A0A0Q9WRL3 A0A182LF97 A0A1A9X7X2 B4I929 A0A182MWJ3
A0A1W6EVY0 A0A182P717 A0A2J7RM97 F4X480 W4VRN2 A0A067RRP6 A0A151I9W8 A0A3L8E7V4 A0A348G601 A0A1V1FVK8 A0A182QQ91 A0A158N9I3 E2AE72 A0A151I452 A0A182VX80 A0A026WDN1 A0A0L7R5M1 A0A182YKF6 A0A2A3EQH4 A0A087ZRN5 A0A0J7KJS1 A0A151JV41 A0A151XGD6 A0A0M3QZA1 A0A182HT57 A0A182NA58 A0A182UMV6 Q7QC23 A0A182XI70 A0A195E1K9 A0A182RI04 E2BNR3 A0A0Q9WMF1 J9HJF8 A0A1W4UVK5 A0A0L0BTV6 A0A1I8Q4W2 A0A182J566 B4PX05 B4MEQ9 B4R7E8 A0A182F709 A0A1Q3G2F0 B3P973 A0A1Q3G2L4 W5JS53 A0A154PFK1 A0A1Q3G2Y6 A0A1Q3G2Y9 A0A1Q3G243 A0A2M4B9D9 P42787-5 A0A1Q3G254 X2J9Z1 A0A2M4B9F1 A0A2M4A298 A0A0P8ZHH2 A0A2M4A2G0 B4L2T3 A0A2M4B9B6 P42787 A0A1W4V999 A0A3B0KTY1 A0A0A1XAK1 A0A1A9UUA2 B4JNA3 B5DMP1 A0A1J1IBI2 A0A0R1EF72 B4GY07 A0A1B6LU96 A0A0K8VVD6 V9IFV8 A0A0Q5SRQ2 A0A1B6BYT4 P42787-4 P42787-3 B4N2H9 B3MYX9 A0A0Q9W7V5 A0A1I8MWK5 A0A2M3YZY6 A0A2M3YZV8 A0A310SMJ0 A0A0R3P055 A0A1B6GCF9 A0A0Q9WRL3 A0A182LF97 A0A1A9X7X2 B4I929 A0A182MWJ3
EC Number
3.4.17.22
Pubmed
19121390
22118469
28004739
21719571
24845553
30249741
+ More
28410430 21347285 20798317 24508170 25244985 12364791 14747013 17210077 17994087 17510324 26108605 17550304 20920257 23761445 12393882 10731132 12537572 10731137 7568156 8000074 17893096 18327897 12537568 12537573 12537574 16110336 17569856 17569867 25830018 15632085 25315136 20966253
28410430 21347285 20798317 24508170 25244985 12364791 14747013 17210077 17994087 17510324 26108605 17550304 20920257 23761445 12393882 10731132 12537572 10731137 7568156 8000074 17893096 18327897 12537568 12537573 12537574 16110336 17569856 17569867 25830018 15632085 25315136 20966253
EMBL
BABH01004042
BABH01004043
RSAL01000071
RVE49138.1
AGBW02007651
OWR55065.1
+ More
GEZM01007557 JAV95093.1 GFDF01005237 JAV08847.1 GFDF01005236 JAV08848.1 KY563465 ARK19874.1 NEVH01002568 PNF41963.1 GL888628 EGI58748.1 GANO01001118 JAB58753.1 KK852507 KDR22449.1 KQ978264 KYM95838.1 QOIP01000001 RLU27578.1 FX985540 BBF97874.1 FX985405 BAX07418.1 AXCN02000215 ADTU01009655 GL438827 EFN68305.1 KQ976461 KYM84710.1 KK107260 EZA54200.1 KQ414648 KOC66182.1 KZ288202 PBC33522.1 LBMM01006597 KMQ90474.1 KQ981713 KYN37346.1 KQ982169 KYQ59464.1 CP012528 ALC49063.1 APCN01000420 AAAB01008859 EAA07478.5 KQ979814 KYN19013.1 GL449444 EFN82672.1 CH933810 KRF93869.1 CH478029 EJY58039.1 JRES01001349 KNC23482.1 CM000162 EDX00791.1 CH940664 EDW63034.2 CM000366 EDX16759.1 GFDL01001037 JAV34008.1 CH954183 EDV45369.2 GFDL01001024 JAV34021.1 ADMH02000331 ETN66931.1 KQ434886 KZC10088.1 GFDL01000919 JAV34126.1 GFDL01000936 JAV34109.1 GFDL01001158 JAV33887.1 GGFJ01000481 MBW49622.1 AF545816 AF545817 AF545818 AF545819 AF545820 AE014298 AL009147 BT029935 BT099720 BT100310 U29591 U29592 U03883 GFDL01001161 JAV33884.1 AHN59215.1 GGFJ01000480 MBW49621.1 GGFK01001615 MBW34936.1 CH902632 KPU74157.1 GGFK01001601 MBW34922.1 EDW06900.1 GGFJ01000482 MBW49623.1 OUUW01000019 SPP89266.1 GBXI01006341 JAD07951.1 CH916371 EDV92196.1 CH379064 EDY72721.1 CVRI01000047 CRK97571.1 KRK05786.1 CH479196 EDW27634.1 GEBQ01012730 JAT27247.1 GDHF01024787 GDHF01009789 JAI27527.1 JAI42525.1 JR040581 AEY59194.1 KQS25945.1 GEDC01031153 JAS06145.1 CH963925 EDW78568.2 EDV32823.2 KRF80877.1 GGFM01001055 MBW21806.1 GGFM01001058 MBW21809.1 KQ759874 OAD62352.1 KRT06602.1 GECZ01009642 JAS60127.1 KRF98900.1 CH480825 EDW43710.1 AXCM01008467
GEZM01007557 JAV95093.1 GFDF01005237 JAV08847.1 GFDF01005236 JAV08848.1 KY563465 ARK19874.1 NEVH01002568 PNF41963.1 GL888628 EGI58748.1 GANO01001118 JAB58753.1 KK852507 KDR22449.1 KQ978264 KYM95838.1 QOIP01000001 RLU27578.1 FX985540 BBF97874.1 FX985405 BAX07418.1 AXCN02000215 ADTU01009655 GL438827 EFN68305.1 KQ976461 KYM84710.1 KK107260 EZA54200.1 KQ414648 KOC66182.1 KZ288202 PBC33522.1 LBMM01006597 KMQ90474.1 KQ981713 KYN37346.1 KQ982169 KYQ59464.1 CP012528 ALC49063.1 APCN01000420 AAAB01008859 EAA07478.5 KQ979814 KYN19013.1 GL449444 EFN82672.1 CH933810 KRF93869.1 CH478029 EJY58039.1 JRES01001349 KNC23482.1 CM000162 EDX00791.1 CH940664 EDW63034.2 CM000366 EDX16759.1 GFDL01001037 JAV34008.1 CH954183 EDV45369.2 GFDL01001024 JAV34021.1 ADMH02000331 ETN66931.1 KQ434886 KZC10088.1 GFDL01000919 JAV34126.1 GFDL01000936 JAV34109.1 GFDL01001158 JAV33887.1 GGFJ01000481 MBW49622.1 AF545816 AF545817 AF545818 AF545819 AF545820 AE014298 AL009147 BT029935 BT099720 BT100310 U29591 U29592 U03883 GFDL01001161 JAV33884.1 AHN59215.1 GGFJ01000480 MBW49621.1 GGFK01001615 MBW34936.1 CH902632 KPU74157.1 GGFK01001601 MBW34922.1 EDW06900.1 GGFJ01000482 MBW49623.1 OUUW01000019 SPP89266.1 GBXI01006341 JAD07951.1 CH916371 EDV92196.1 CH379064 EDY72721.1 CVRI01000047 CRK97571.1 KRK05786.1 CH479196 EDW27634.1 GEBQ01012730 JAT27247.1 GDHF01024787 GDHF01009789 JAI27527.1 JAI42525.1 JR040581 AEY59194.1 KQS25945.1 GEDC01031153 JAS06145.1 CH963925 EDW78568.2 EDV32823.2 KRF80877.1 GGFM01001055 MBW21806.1 GGFM01001058 MBW21809.1 KQ759874 OAD62352.1 KRT06602.1 GECZ01009642 JAS60127.1 KRF98900.1 CH480825 EDW43710.1 AXCM01008467
Proteomes
UP000005204
UP000283053
UP000007151
UP000075885
UP000235965
UP000007755
+ More
UP000027135 UP000078542 UP000279307 UP000075886 UP000005205 UP000000311 UP000078540 UP000075920 UP000053097 UP000053825 UP000076408 UP000242457 UP000005203 UP000036403 UP000078541 UP000075809 UP000092553 UP000075840 UP000075884 UP000075903 UP000007062 UP000076407 UP000078492 UP000075900 UP000008237 UP000009192 UP000008820 UP000192221 UP000037069 UP000095300 UP000075880 UP000002282 UP000008792 UP000000304 UP000069272 UP000008711 UP000000673 UP000076502 UP000000803 UP000007801 UP000268350 UP000078200 UP000001070 UP000001819 UP000183832 UP000008744 UP000007798 UP000095301 UP000075882 UP000092443 UP000001292 UP000075883
UP000027135 UP000078542 UP000279307 UP000075886 UP000005205 UP000000311 UP000078540 UP000075920 UP000053097 UP000053825 UP000076408 UP000242457 UP000005203 UP000036403 UP000078541 UP000075809 UP000092553 UP000075840 UP000075884 UP000075903 UP000007062 UP000076407 UP000078492 UP000075900 UP000008237 UP000009192 UP000008820 UP000192221 UP000037069 UP000095300 UP000075880 UP000002282 UP000008792 UP000000304 UP000069272 UP000008711 UP000000673 UP000076502 UP000000803 UP000007801 UP000268350 UP000078200 UP000001070 UP000001819 UP000183832 UP000008744 UP000007798 UP000095301 UP000075882 UP000092443 UP000001292 UP000075883
Pfam
PF00246 Peptidase_M14
Interpro
SUPFAM
SSF49464
SSF49464
ProteinModelPortal
H9JTE5
A0A3S2M1L9
A0A212FMU7
A0A1Y1NB52
A0A1L8DR43
A0A1L8DQU3
+ More
A0A1W6EVY0 A0A182P717 A0A2J7RM97 F4X480 W4VRN2 A0A067RRP6 A0A151I9W8 A0A3L8E7V4 A0A348G601 A0A1V1FVK8 A0A182QQ91 A0A158N9I3 E2AE72 A0A151I452 A0A182VX80 A0A026WDN1 A0A0L7R5M1 A0A182YKF6 A0A2A3EQH4 A0A087ZRN5 A0A0J7KJS1 A0A151JV41 A0A151XGD6 A0A0M3QZA1 A0A182HT57 A0A182NA58 A0A182UMV6 Q7QC23 A0A182XI70 A0A195E1K9 A0A182RI04 E2BNR3 A0A0Q9WMF1 J9HJF8 A0A1W4UVK5 A0A0L0BTV6 A0A1I8Q4W2 A0A182J566 B4PX05 B4MEQ9 B4R7E8 A0A182F709 A0A1Q3G2F0 B3P973 A0A1Q3G2L4 W5JS53 A0A154PFK1 A0A1Q3G2Y6 A0A1Q3G2Y9 A0A1Q3G243 A0A2M4B9D9 P42787-5 A0A1Q3G254 X2J9Z1 A0A2M4B9F1 A0A2M4A298 A0A0P8ZHH2 A0A2M4A2G0 B4L2T3 A0A2M4B9B6 P42787 A0A1W4V999 A0A3B0KTY1 A0A0A1XAK1 A0A1A9UUA2 B4JNA3 B5DMP1 A0A1J1IBI2 A0A0R1EF72 B4GY07 A0A1B6LU96 A0A0K8VVD6 V9IFV8 A0A0Q5SRQ2 A0A1B6BYT4 P42787-4 P42787-3 B4N2H9 B3MYX9 A0A0Q9W7V5 A0A1I8MWK5 A0A2M3YZY6 A0A2M3YZV8 A0A310SMJ0 A0A0R3P055 A0A1B6GCF9 A0A0Q9WRL3 A0A182LF97 A0A1A9X7X2 B4I929 A0A182MWJ3
A0A1W6EVY0 A0A182P717 A0A2J7RM97 F4X480 W4VRN2 A0A067RRP6 A0A151I9W8 A0A3L8E7V4 A0A348G601 A0A1V1FVK8 A0A182QQ91 A0A158N9I3 E2AE72 A0A151I452 A0A182VX80 A0A026WDN1 A0A0L7R5M1 A0A182YKF6 A0A2A3EQH4 A0A087ZRN5 A0A0J7KJS1 A0A151JV41 A0A151XGD6 A0A0M3QZA1 A0A182HT57 A0A182NA58 A0A182UMV6 Q7QC23 A0A182XI70 A0A195E1K9 A0A182RI04 E2BNR3 A0A0Q9WMF1 J9HJF8 A0A1W4UVK5 A0A0L0BTV6 A0A1I8Q4W2 A0A182J566 B4PX05 B4MEQ9 B4R7E8 A0A182F709 A0A1Q3G2F0 B3P973 A0A1Q3G2L4 W5JS53 A0A154PFK1 A0A1Q3G2Y6 A0A1Q3G2Y9 A0A1Q3G243 A0A2M4B9D9 P42787-5 A0A1Q3G254 X2J9Z1 A0A2M4B9F1 A0A2M4A298 A0A0P8ZHH2 A0A2M4A2G0 B4L2T3 A0A2M4B9B6 P42787 A0A1W4V999 A0A3B0KTY1 A0A0A1XAK1 A0A1A9UUA2 B4JNA3 B5DMP1 A0A1J1IBI2 A0A0R1EF72 B4GY07 A0A1B6LU96 A0A0K8VVD6 V9IFV8 A0A0Q5SRQ2 A0A1B6BYT4 P42787-4 P42787-3 B4N2H9 B3MYX9 A0A0Q9W7V5 A0A1I8MWK5 A0A2M3YZY6 A0A2M3YZV8 A0A310SMJ0 A0A0R3P055 A0A1B6GCF9 A0A0Q9WRL3 A0A182LF97 A0A1A9X7X2 B4I929 A0A182MWJ3
PDB
3MN8
E-value=9.42672e-115,
Score=1063
Ontologies
GO
PANTHER
Topology
Subcellular location
Membrane
SignalP
Position: 1 - 19,
Likelihood: 0.997669
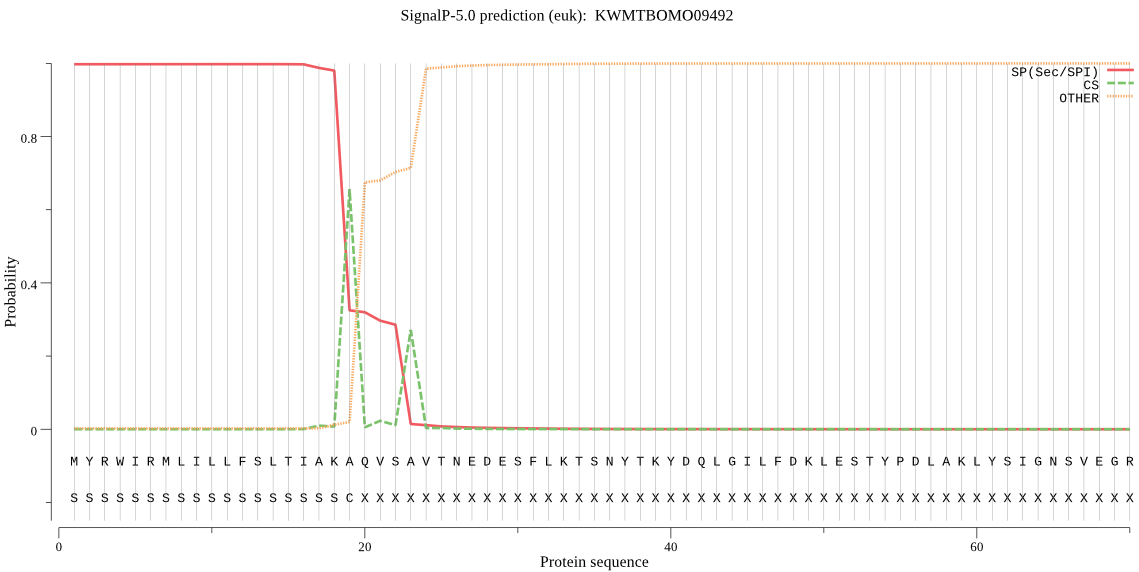
Length:
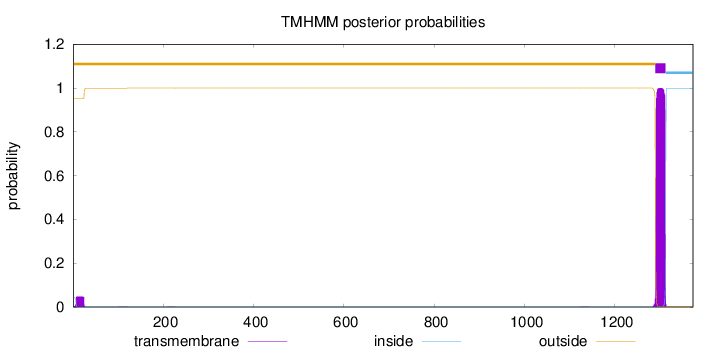
1373
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.58262
Exp number, first 60 AAs:
0.89644
Total prob of N-in:
0.04760
outside
1 - 1289
TMhelix
1290 - 1312
inside
1313 - 1373
Population Genetic Test Statistics
Pi
225.566724
Theta
176.771585
Tajima's D
0.731334
CLR
0.338524
CSRT
0.583220838958052
Interpretation
Uncertain
