Gene
KWMTBOMO09481
Pre Gene Modal
BGIBMGA012801
Annotation
CRAL-TRIO_domain-containing_protein?_partial_[Manduca_sexta]
Location in the cell
Mitochondrial Reliability : 2.464
Sequence
CDS
ATGCGCGGTGTACGTTTTGGACACATAACGAGAATTAGTTTATTGTCGCTGCGTAAATTCTTCAATTACATTCAGGAAGGCGTCCCGGTCCGTATGCGGGCTATCCACGTTCTGAACACCGAACCCGTTTTGGATAAACTCATATTGTTGATCAGGCCGTTTATGGACAAAAAGTTTTTTGATATGATAAAATTCCACAACAAAACCGAAGACCTGGAATCGTTTTACGAAACGGCAATACCCAGGTCGTCCCTGCCACCGGACCACGGAGGAACGCTTCCCGACACCCAGACTCTGCACAAGAAATGCATGCAGCAACTCCAGCTCCTAGAGCCGTACTTCAAGGCTGAAGAGGAACAGAGAATCGCCGCTCTGCCCGACAAGAAACGGGAGAAGGCGATGGAGAAAGCCTTCAAGAATCTAGAGATAGATTAA
Protein
MRGVRFGHITRISLLSLRKFFNYIQEGVPVRMRAIHVLNTEPVLDKLILLIRPFMDKKFFDMIKFHNKTEDLESFYETAIPRSSLPPDHGGTLPDTQTLHKKCMQQLQLLEPYFKAEEEQRIAALPDKKREKAMEKAFKNLEID
Summary
Uniprot
A0A2W1BZD3
H9JTD9
A0A0S2Z353
A0A2A4JYQ6
A0A194Q0E0
A0A2H4RMT5
+ More
A0A2H4RMT0 A0A1E1WUJ1 A0A212FMV7 A0A0L7LTE7 A0A437BBU4 A0A182R5Q7 A0A1B6JB60 A0A1B6ISW4 A0A182YP36 A0A182QMV7 A0A182PKF9 A0A182M2D4 A0A1L8DBU5 A0A1B6MQ64 A0A1L8DC92 A0A1B6MKT4 A0A182VY12 A0A2S2PVS5 A0A182UD92 A0A182N4K4 A0A182LAY9 A0A182WVC4 Q7QG16 A0A182I905 A0A182K9N6 A0A182US03 B0W3K4 A0A1Q3FJ52 A0A1B0CL61 A0A084VZX2 A0A1B6GXD7 A0A182J163 A0A023F815 W5JCQ4 A0A182H7H9 A0A023ENS0 A0A2M4AY24 Q16RH6 A0A182F946 A0A2M4BUV4 A0A2M4BV73 A0A2M4BV13 A0A2M4BVA0 A0A2H8TEU9 Q1HRG7 A0A1B6DDH3 A0A2R7WSN1 J9JP41 A0A1J1IPD0 K7IS61 A0A232F498 A0A170X858 A0A2S2QHT7 R4G355 A0A1S3DP78 A0A0C9RFG4 A0A336K1C6 A0A224XHE8 A0A2S2NPS6 E9IA70 A0A2M4BWV0 A0A139WGJ3 A0A0A9YEI6 A0A0A9YG14 A0A0K8TCH4 A0A146KP30 E2C418 A0A0A9YHJ0 A0A0K8TDH4 A0A067R4X7 A0A1B6E4Z5 A0A1B6D0A6 A0A026W2Z0 A0A3L8DTS9 E2BL70 A0A139WDA0 A0A2P8XNT8 A0A2J7PS36 A0A1B6KMT6 A0A195FRC0 D2A504 A0A195ECZ2 E2ADJ9 A0A1B6GM42 A0A139WD63 U4UZ61 A0A0J7KK55 A0A0J7NH54 A0A1Y1JX72 A0A1Y1JWP1 E9IJH9 N6TBB2 A0A1Y1LY65
A0A2H4RMT0 A0A1E1WUJ1 A0A212FMV7 A0A0L7LTE7 A0A437BBU4 A0A182R5Q7 A0A1B6JB60 A0A1B6ISW4 A0A182YP36 A0A182QMV7 A0A182PKF9 A0A182M2D4 A0A1L8DBU5 A0A1B6MQ64 A0A1L8DC92 A0A1B6MKT4 A0A182VY12 A0A2S2PVS5 A0A182UD92 A0A182N4K4 A0A182LAY9 A0A182WVC4 Q7QG16 A0A182I905 A0A182K9N6 A0A182US03 B0W3K4 A0A1Q3FJ52 A0A1B0CL61 A0A084VZX2 A0A1B6GXD7 A0A182J163 A0A023F815 W5JCQ4 A0A182H7H9 A0A023ENS0 A0A2M4AY24 Q16RH6 A0A182F946 A0A2M4BUV4 A0A2M4BV73 A0A2M4BV13 A0A2M4BVA0 A0A2H8TEU9 Q1HRG7 A0A1B6DDH3 A0A2R7WSN1 J9JP41 A0A1J1IPD0 K7IS61 A0A232F498 A0A170X858 A0A2S2QHT7 R4G355 A0A1S3DP78 A0A0C9RFG4 A0A336K1C6 A0A224XHE8 A0A2S2NPS6 E9IA70 A0A2M4BWV0 A0A139WGJ3 A0A0A9YEI6 A0A0A9YG14 A0A0K8TCH4 A0A146KP30 E2C418 A0A0A9YHJ0 A0A0K8TDH4 A0A067R4X7 A0A1B6E4Z5 A0A1B6D0A6 A0A026W2Z0 A0A3L8DTS9 E2BL70 A0A139WDA0 A0A2P8XNT8 A0A2J7PS36 A0A1B6KMT6 A0A195FRC0 D2A504 A0A195ECZ2 E2ADJ9 A0A1B6GM42 A0A139WD63 U4UZ61 A0A0J7KK55 A0A0J7NH54 A0A1Y1JX72 A0A1Y1JWP1 E9IJH9 N6TBB2 A0A1Y1LY65
Pubmed
28756777
19121390
25684408
26354079
29136137
22118469
+ More
26227816 25244985 20966253 12364791 14747013 17210077 24438588 25474469 20920257 23761445 26483478 24945155 17510324 17204158 20075255 28648823 21282665 18362917 19820115 25401762 26823975 20798317 24845553 24508170 30249741 29403074 23537049 28004739
26227816 25244985 20966253 12364791 14747013 17210077 24438588 25474469 20920257 23761445 26483478 24945155 17510324 17204158 20075255 28648823 21282665 18362917 19820115 25401762 26823975 20798317 24845553 24508170 30249741 29403074 23537049 28004739
EMBL
KZ149935
PZC77173.1
BABH01004068
KT943560
ALQ33318.1
NWSH01000386
+ More
PCG76774.1 KQ459582 KPI99047.1 MG434647 ATY51953.1 MG434636 ATY51942.1 GDQN01000406 JAT90648.1 AGBW02007651 OWR55075.1 JTDY01000160 KOB78516.1 RSAL01000093 RVE47886.1 GECU01023166 GECU01011302 JAS84540.1 JAS96404.1 GECU01017682 JAS90024.1 AXCN02000044 AXCM01004998 GFDF01010151 JAV03933.1 GEBQ01001890 JAT38087.1 GFDF01010150 JAV03934.1 GEBQ01003498 JAT36479.1 GGMR01020347 MBY32966.1 AAAB01008844 EAA05957.5 EGK96341.1 APCN01003147 DS231832 EDS31790.1 GFDL01007532 JAV27513.1 AJWK01016949 AJWK01016950 AJWK01016951 AJWK01016952 ATLV01019013 KE525259 KFB43516.1 GECZ01002683 JAS67086.1 GBBI01001305 JAC17407.1 ADMH02001893 ETN60609.1 JXUM01028270 JXUM01028271 JXUM01028272 JXUM01028273 GAPW01003012 JAC10586.1 GGFK01012359 MBW45680.1 CH477709 EAT37013.1 GGFJ01007719 MBW56860.1 GGFJ01007818 MBW56959.1 GGFJ01007467 MBW56608.1 GGFJ01007720 MBW56861.1 GFXV01000427 MBW12232.1 DQ440127 ABF18160.1 GEDC01013602 JAS23696.1 KK855438 PTY22572.1 ABLF02018999 ABLF02019001 ABLF02046099 CVRI01000057 CRL02024.1 NNAY01001062 OXU25290.1 GEMB01004696 JAR98592.1 GGMS01008081 MBY77284.1 ACPB03022651 GAHY01001986 JAA75524.1 GBYB01015329 GBYB01015330 JAG85096.1 JAG85097.1 UFQS01000022 UFQT01000022 SSW97612.1 SSX17998.1 GFTR01004510 JAW11916.1 GGMR01006562 MBY19181.1 GL761987 EFZ22528.1 GGFJ01008424 MBW57565.1 KQ971348 KYB26927.1 GBHO01013039 GBHO01013035 JAG30565.1 JAG30569.1 GBHO01013034 GBHO01013033 JAG30570.1 JAG30571.1 GBRD01002607 JAG63214.1 GDHC01021597 GDHC01008315 GDHC01002734 JAP97031.1 JAQ10314.1 JAQ15895.1 GL452364 EFN77404.1 GBHO01013038 GBHO01013036 JAG30566.1 JAG30568.1 GBRD01002606 JAG63215.1 KK852693 KDR18315.1 GEDC01004295 JAS33003.1 GEDC01018207 JAS19091.1 KK107459 EZA50388.1 QOIP01000004 RLU23712.1 GL448949 EFN83556.1 KQ971361 KYB25884.1 PYGN01001631 PSN33670.1 NEVH01021939 PNF19141.1 GEBQ01027209 JAT12768.1 KQ981305 KYN42976.1 EFA05290.2 KQ979074 KYN23073.1 GL438812 EFN68493.1 GECZ01006271 GECZ01004148 JAS63498.1 JAS65621.1 KYB25883.1 KI207135 ERL95635.1 LBMM01006207 KMQ90808.1 LBMM01005052 KMQ91855.1 GEZM01098820 JAV53733.1 GEZM01098819 JAV53734.1 GL763802 EFZ19284.1 APGK01044181 KB741021 ENN75018.1 GEZM01044921 JAV78041.1
PCG76774.1 KQ459582 KPI99047.1 MG434647 ATY51953.1 MG434636 ATY51942.1 GDQN01000406 JAT90648.1 AGBW02007651 OWR55075.1 JTDY01000160 KOB78516.1 RSAL01000093 RVE47886.1 GECU01023166 GECU01011302 JAS84540.1 JAS96404.1 GECU01017682 JAS90024.1 AXCN02000044 AXCM01004998 GFDF01010151 JAV03933.1 GEBQ01001890 JAT38087.1 GFDF01010150 JAV03934.1 GEBQ01003498 JAT36479.1 GGMR01020347 MBY32966.1 AAAB01008844 EAA05957.5 EGK96341.1 APCN01003147 DS231832 EDS31790.1 GFDL01007532 JAV27513.1 AJWK01016949 AJWK01016950 AJWK01016951 AJWK01016952 ATLV01019013 KE525259 KFB43516.1 GECZ01002683 JAS67086.1 GBBI01001305 JAC17407.1 ADMH02001893 ETN60609.1 JXUM01028270 JXUM01028271 JXUM01028272 JXUM01028273 GAPW01003012 JAC10586.1 GGFK01012359 MBW45680.1 CH477709 EAT37013.1 GGFJ01007719 MBW56860.1 GGFJ01007818 MBW56959.1 GGFJ01007467 MBW56608.1 GGFJ01007720 MBW56861.1 GFXV01000427 MBW12232.1 DQ440127 ABF18160.1 GEDC01013602 JAS23696.1 KK855438 PTY22572.1 ABLF02018999 ABLF02019001 ABLF02046099 CVRI01000057 CRL02024.1 NNAY01001062 OXU25290.1 GEMB01004696 JAR98592.1 GGMS01008081 MBY77284.1 ACPB03022651 GAHY01001986 JAA75524.1 GBYB01015329 GBYB01015330 JAG85096.1 JAG85097.1 UFQS01000022 UFQT01000022 SSW97612.1 SSX17998.1 GFTR01004510 JAW11916.1 GGMR01006562 MBY19181.1 GL761987 EFZ22528.1 GGFJ01008424 MBW57565.1 KQ971348 KYB26927.1 GBHO01013039 GBHO01013035 JAG30565.1 JAG30569.1 GBHO01013034 GBHO01013033 JAG30570.1 JAG30571.1 GBRD01002607 JAG63214.1 GDHC01021597 GDHC01008315 GDHC01002734 JAP97031.1 JAQ10314.1 JAQ15895.1 GL452364 EFN77404.1 GBHO01013038 GBHO01013036 JAG30566.1 JAG30568.1 GBRD01002606 JAG63215.1 KK852693 KDR18315.1 GEDC01004295 JAS33003.1 GEDC01018207 JAS19091.1 KK107459 EZA50388.1 QOIP01000004 RLU23712.1 GL448949 EFN83556.1 KQ971361 KYB25884.1 PYGN01001631 PSN33670.1 NEVH01021939 PNF19141.1 GEBQ01027209 JAT12768.1 KQ981305 KYN42976.1 EFA05290.2 KQ979074 KYN23073.1 GL438812 EFN68493.1 GECZ01006271 GECZ01004148 JAS63498.1 JAS65621.1 KYB25883.1 KI207135 ERL95635.1 LBMM01006207 KMQ90808.1 LBMM01005052 KMQ91855.1 GEZM01098820 JAV53733.1 GEZM01098819 JAV53734.1 GL763802 EFZ19284.1 APGK01044181 KB741021 ENN75018.1 GEZM01044921 JAV78041.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000037510
UP000283053
+ More
UP000075900 UP000076408 UP000075886 UP000075885 UP000075883 UP000075920 UP000075902 UP000075884 UP000075882 UP000076407 UP000007062 UP000075840 UP000075881 UP000075903 UP000002320 UP000092461 UP000030765 UP000075880 UP000000673 UP000069940 UP000008820 UP000069272 UP000007819 UP000183832 UP000002358 UP000215335 UP000015103 UP000079169 UP000007266 UP000008237 UP000027135 UP000053097 UP000279307 UP000245037 UP000235965 UP000078541 UP000078492 UP000000311 UP000030742 UP000036403 UP000019118
UP000075900 UP000076408 UP000075886 UP000075885 UP000075883 UP000075920 UP000075902 UP000075884 UP000075882 UP000076407 UP000007062 UP000075840 UP000075881 UP000075903 UP000002320 UP000092461 UP000030765 UP000075880 UP000000673 UP000069940 UP000008820 UP000069272 UP000007819 UP000183832 UP000002358 UP000215335 UP000015103 UP000079169 UP000007266 UP000008237 UP000027135 UP000053097 UP000279307 UP000245037 UP000235965 UP000078541 UP000078492 UP000000311 UP000030742 UP000036403 UP000019118
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A2W1BZD3
H9JTD9
A0A0S2Z353
A0A2A4JYQ6
A0A194Q0E0
A0A2H4RMT5
+ More
A0A2H4RMT0 A0A1E1WUJ1 A0A212FMV7 A0A0L7LTE7 A0A437BBU4 A0A182R5Q7 A0A1B6JB60 A0A1B6ISW4 A0A182YP36 A0A182QMV7 A0A182PKF9 A0A182M2D4 A0A1L8DBU5 A0A1B6MQ64 A0A1L8DC92 A0A1B6MKT4 A0A182VY12 A0A2S2PVS5 A0A182UD92 A0A182N4K4 A0A182LAY9 A0A182WVC4 Q7QG16 A0A182I905 A0A182K9N6 A0A182US03 B0W3K4 A0A1Q3FJ52 A0A1B0CL61 A0A084VZX2 A0A1B6GXD7 A0A182J163 A0A023F815 W5JCQ4 A0A182H7H9 A0A023ENS0 A0A2M4AY24 Q16RH6 A0A182F946 A0A2M4BUV4 A0A2M4BV73 A0A2M4BV13 A0A2M4BVA0 A0A2H8TEU9 Q1HRG7 A0A1B6DDH3 A0A2R7WSN1 J9JP41 A0A1J1IPD0 K7IS61 A0A232F498 A0A170X858 A0A2S2QHT7 R4G355 A0A1S3DP78 A0A0C9RFG4 A0A336K1C6 A0A224XHE8 A0A2S2NPS6 E9IA70 A0A2M4BWV0 A0A139WGJ3 A0A0A9YEI6 A0A0A9YG14 A0A0K8TCH4 A0A146KP30 E2C418 A0A0A9YHJ0 A0A0K8TDH4 A0A067R4X7 A0A1B6E4Z5 A0A1B6D0A6 A0A026W2Z0 A0A3L8DTS9 E2BL70 A0A139WDA0 A0A2P8XNT8 A0A2J7PS36 A0A1B6KMT6 A0A195FRC0 D2A504 A0A195ECZ2 E2ADJ9 A0A1B6GM42 A0A139WD63 U4UZ61 A0A0J7KK55 A0A0J7NH54 A0A1Y1JX72 A0A1Y1JWP1 E9IJH9 N6TBB2 A0A1Y1LY65
A0A2H4RMT0 A0A1E1WUJ1 A0A212FMV7 A0A0L7LTE7 A0A437BBU4 A0A182R5Q7 A0A1B6JB60 A0A1B6ISW4 A0A182YP36 A0A182QMV7 A0A182PKF9 A0A182M2D4 A0A1L8DBU5 A0A1B6MQ64 A0A1L8DC92 A0A1B6MKT4 A0A182VY12 A0A2S2PVS5 A0A182UD92 A0A182N4K4 A0A182LAY9 A0A182WVC4 Q7QG16 A0A182I905 A0A182K9N6 A0A182US03 B0W3K4 A0A1Q3FJ52 A0A1B0CL61 A0A084VZX2 A0A1B6GXD7 A0A182J163 A0A023F815 W5JCQ4 A0A182H7H9 A0A023ENS0 A0A2M4AY24 Q16RH6 A0A182F946 A0A2M4BUV4 A0A2M4BV73 A0A2M4BV13 A0A2M4BVA0 A0A2H8TEU9 Q1HRG7 A0A1B6DDH3 A0A2R7WSN1 J9JP41 A0A1J1IPD0 K7IS61 A0A232F498 A0A170X858 A0A2S2QHT7 R4G355 A0A1S3DP78 A0A0C9RFG4 A0A336K1C6 A0A224XHE8 A0A2S2NPS6 E9IA70 A0A2M4BWV0 A0A139WGJ3 A0A0A9YEI6 A0A0A9YG14 A0A0K8TCH4 A0A146KP30 E2C418 A0A0A9YHJ0 A0A0K8TDH4 A0A067R4X7 A0A1B6E4Z5 A0A1B6D0A6 A0A026W2Z0 A0A3L8DTS9 E2BL70 A0A139WDA0 A0A2P8XNT8 A0A2J7PS36 A0A1B6KMT6 A0A195FRC0 D2A504 A0A195ECZ2 E2ADJ9 A0A1B6GM42 A0A139WD63 U4UZ61 A0A0J7KK55 A0A0J7NH54 A0A1Y1JX72 A0A1Y1JWP1 E9IJH9 N6TBB2 A0A1Y1LY65
PDB
3HY5
E-value=0.00170996,
Score=92
Ontologies
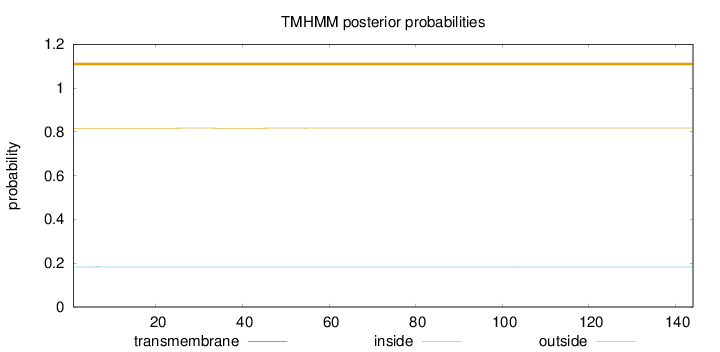
Topology
Length:
144
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01851
Exp number, first 60 AAs:
0.01851
Total prob of N-in:
0.18309
outside
1 - 144
Population Genetic Test Statistics
Pi
235.553656
Theta
169.294523
Tajima's D
1.318482
CLR
0.013322
CSRT
0.743062846857657
Interpretation
Uncertain
