Gene
KWMTBOMO09479
Pre Gene Modal
BGIBMGA013061
Annotation
PREDICTED:_GPI_transamidase_component_PIG-S_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.706
Sequence
CDS
ATGTGGGCCTCTGCTTCATTCGTAGGGGTCCTAATCATCATCGGGATGCCGCTATGGTGGAAAACTACGGAAGTCTACAGGGTGTCGCTGCCGTACGAAAAGATTTCATCGTTCAAACCTCTATCGCATTTCGTCACAACTGAACTGAAAGTCTTAGCCAACGATGACGCTACAGCATCTAAGATCGTCAGCGAGATACAGAAGGCATTTGCTGAATCTGATATAATAAAGATCAAAATCGAGAAAACAGTAATGTCGGAGCGACAACAGCACATTTTGCGGTCGGTGGTTGATATGGAGAAAGCCGTCGAAGAACTAGCTTCTACATTGGACCTCGACCAAGAAAATATATTCCACGTGGTGCAAAAGAAATCGTTGTCTCACAATATCTGGGTTGGCAACGAAAGAATAATGTTCTTCGATGATGAGAAAGCATCAGAAACGATAGTGAAAGCGTTAGACGGCTGGATATATCAGACTTCGGTACTCCGAGGATCCAGATCGGACACTCCTGATCAAGCCCGAAGCACGAGGTTCCCGCCGGAGTCGGCCTACCACATCACACTGTCCATAGTGCATCCAAGGCCTGATACAATGAAAGTTGTTTGGAATGCCGCTGAAGCTGTCGAGGATTACATTGGCAGCTTTGTCGACGAGTTAAGCTCCTTGCACAACTTCACGCTGAAGTCGCAATGGCTTCATCTACTCGACTTCGACTTTCAAGCTAGAGAGGTAAAAGAAGTCAGTGAATGGGGACGTCACTTCGCCGTGCGTCAGGACAGGCTGTCATTGTTATTGACAAGGCTCGAGGAACGAGCTGCGACGCACGTCTCTGAGCTATCAACGATCAACCTGGCGCTCTACATCGTTCCCTGCGATAAAGCTCCCATCATTATATACGATAATGAAGGTCGCAAAAATCCTTTGTCGATACAAGCATTTATGTCCCCCAAGTGGGGGGGCGTGGTGATCGCCAGTCCCCCGGCCGAGCAGTGCTCCCAGCAGGTCCGAGAGTTCGAGCCCAGCGTGGGACAGATCATGGGCGCGTTCGTGTCCCACCTCAGGGAGTTGCTCGGGATCACGAAAAAGGACGCAATCGAAGGAGCACACTTAGAGCCTTCGAGGTCGGTGTCGCCGCGCCGCTGGGAGGTGGACGCGCTACTCCGCATCAGGACCTTAGAGCAGGTCACATCCGCTGAAAGAACTCTACAGTCTCTAGCGCAATTATTGGAGGAGATATCGAACATAGTGATAAACGATGAAGTGGGCGCGTCCATCGCGGCCGCGGTGTCCGAGCTGGAGGCGGGCGCGGGGGAGGGCGCGGGGGCGGACGCGGCGCGGGCGGGGCGGGCGCACGCGCATGCGCAGCGCGCCTTCACGCACCCCGGTCTACTCGCACTGCTGTACTTCCCGGACGATCAGAAATACGCGATCTACATACCGTTGTTTTTACCGATAATGTTCCCGGTGGTTTTGTCATTGAGAATATTGTTATTATGGTTCATGGGGAAGCCGGTGCACAAAGAAAAAACTGATTGA
Protein
MWASASFVGVLIIIGMPLWWKTTEVYRVSLPYEKISSFKPLSHFVTTELKVLANDDATASKIVSEIQKAFAESDIIKIKIEKTVMSERQQHILRSVVDMEKAVEELASTLDLDQENIFHVVQKKSLSHNIWVGNERIMFFDDEKASETIVKALDGWIYQTSVLRGSRSDTPDQARSTRFPPESAYHITLSIVHPRPDTMKVVWNAAEAVEDYIGSFVDELSSLHNFTLKSQWLHLLDFDFQAREVKEVSEWGRHFAVRQDRLSLLLTRLEERAATHVSELSTINLALYIVPCDKAPIIIYDNEGRKNPLSIQAFMSPKWGGVVIASPPAEQCSQQVREFEPSVGQIMGAFVSHLRELLGITKKDAIEGAHLEPSRSVSPRRWEVDALLRIRTLEQVTSAERTLQSLAQLLEEISNIVINDEVGASIAAAVSELEAGAGEGAGADAARAGRAHAHAQRAFTHPGLLALLYFPDDQKYAIYIPLFLPIMFPVVLSLRILLLWFMGKPVHKEKTD
Summary
Uniprot
A0A2H1VAI5
A0A2A4JFF9
A0A3S2PD05
A0A194Q220
A0A212FMZ7
A0A0N0PC78
+ More
A0A2H1WHA7 A0A087ZP46 A0A2A3EPX6 F4W7J1 A0A195EL23 A0A158NPY5 A0A151X8H3 A0A195ESL5 A0A0N0U5B5 E9IWW3 A0A2A3ENP4 E2AWA0 A0A2J7QAS0 A0A2J7QAQ0 A0A2J7QAR0 A0A154PRQ6 V9I7U5 A0A0J7KT94 A0A195ATP6 A0A067QRZ5 A0A0L7RA97 W8BSR2 A0A026WU23 E2BYK5 A0A0K8U5I9 A0A034WBH5 A0A1B6LI78 A0A0A1X6D4 K7IW73 A0A232FE10 A0A1I8JW47 A0A1B6GFU4 Q7QD35 A0A336KT59 A0A1I8PFA1 A0A182Q707 W5JK02 A0A2M4CGM8 A0A2M4CG41 Q171Z3 A0A2M4BIC2 D3TNH7 A0A2M4BID8 A0A182UY32 E0VVQ2 A0A1I8M0S5 A0A2M4AK84 A0A182TQJ0 A0A182LG37 A0A0L0CQJ2 A0A182M739 H6WAR0 A0A1B0BTU3 A0A182FNT1 A0A1A9XDK3 A0A182VRR3 A0A182XV12 A0A1W4VKY4 A0A1B0CEZ4 A0A2P8XP31 A0A182GSS5 A0A1A9ZGD9 A0A182GU49 A0A182JXQ6 B4PV15 B4QU73 A0A0N8P1P5 Q9VC10 U5EV01 B3P6V6 B8A3W8 A0A1L8E3R1 A0A182NLH8 Q86NX1 B5DXD5 B4M653 B4JYA3 T1HSQ2 A0A3L8DAA6 A0A224XLJ6 R4FKN9 A0A0V0G8U0 A0A3B0JD79 A0A182HTZ1 B4G3K7 A0A1J1J3M9 A0A1A9WRR7 B4KBS4 B4NI44 A0A0M4EUJ5 A0A1A9VTW3
A0A2H1WHA7 A0A087ZP46 A0A2A3EPX6 F4W7J1 A0A195EL23 A0A158NPY5 A0A151X8H3 A0A195ESL5 A0A0N0U5B5 E9IWW3 A0A2A3ENP4 E2AWA0 A0A2J7QAS0 A0A2J7QAQ0 A0A2J7QAR0 A0A154PRQ6 V9I7U5 A0A0J7KT94 A0A195ATP6 A0A067QRZ5 A0A0L7RA97 W8BSR2 A0A026WU23 E2BYK5 A0A0K8U5I9 A0A034WBH5 A0A1B6LI78 A0A0A1X6D4 K7IW73 A0A232FE10 A0A1I8JW47 A0A1B6GFU4 Q7QD35 A0A336KT59 A0A1I8PFA1 A0A182Q707 W5JK02 A0A2M4CGM8 A0A2M4CG41 Q171Z3 A0A2M4BIC2 D3TNH7 A0A2M4BID8 A0A182UY32 E0VVQ2 A0A1I8M0S5 A0A2M4AK84 A0A182TQJ0 A0A182LG37 A0A0L0CQJ2 A0A182M739 H6WAR0 A0A1B0BTU3 A0A182FNT1 A0A1A9XDK3 A0A182VRR3 A0A182XV12 A0A1W4VKY4 A0A1B0CEZ4 A0A2P8XP31 A0A182GSS5 A0A1A9ZGD9 A0A182GU49 A0A182JXQ6 B4PV15 B4QU73 A0A0N8P1P5 Q9VC10 U5EV01 B3P6V6 B8A3W8 A0A1L8E3R1 A0A182NLH8 Q86NX1 B5DXD5 B4M653 B4JYA3 T1HSQ2 A0A3L8DAA6 A0A224XLJ6 R4FKN9 A0A0V0G8U0 A0A3B0JD79 A0A182HTZ1 B4G3K7 A0A1J1J3M9 A0A1A9WRR7 B4KBS4 B4NI44 A0A0M4EUJ5 A0A1A9VTW3
Pubmed
26354079
22118469
21719571
21347285
21282665
20798317
+ More
24845553 24495485 24508170 25348373 25830018 20075255 28648823 12364791 14747013 17210077 20920257 23761445 17510324 20353571 20566863 25315136 20966253 26108605 23055932 25244985 29403074 26483478 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 30249741
24845553 24495485 24508170 25348373 25830018 20075255 28648823 12364791 14747013 17210077 20920257 23761445 17510324 20353571 20566863 25315136 20966253 26108605 23055932 25244985 29403074 26483478 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 30249741
EMBL
ODYU01001508
SOQ37811.1
NWSH01001675
PCG70448.1
RSAL01000093
RVE47889.1
+ More
KQ459582 KPI99044.1 AGBW02007651 OWR55079.1 KQ460651 KPJ13034.1 ODYU01008673 SOQ52455.1 KZ288204 PBC33322.1 GL887844 EGI69864.1 KQ978730 KYN28955.1 ADTU01022700 ADTU01022701 ADTU01022702 KQ982409 KYQ56676.1 KQ981986 KYN31240.1 KQ435784 KOX74552.1 GL766616 EFZ14930.1 PBC33320.1 GL443269 EFN62290.1 NEVH01016329 PNF25674.1 PNF25670.1 PNF25673.1 KQ435090 KZC14605.1 JR036511 JR036513 AEY57188.1 AEY57190.1 LBMM01003497 KMQ93464.1 KQ976745 KYM75427.1 KK853383 KDR07999.1 KQ414619 KOC67769.1 GAMC01004368 JAC02188.1 KK107105 EZA59527.1 GL451493 EFN79210.1 GDHF01030674 GDHF01002354 JAI21640.1 JAI49960.1 GAKP01007457 GAKP01007456 JAC51495.1 GEBQ01016594 JAT23383.1 GBXI01007433 JAD06859.1 NNAY01000414 OXU28577.1 GECZ01008465 JAS61304.1 AAAB01008859 EAA07616.5 UFQS01000993 UFQT01000993 SSX08310.1 SSX28333.1 AXCN02000138 ADMH02001019 ETN64436.1 GGFL01000133 MBW64311.1 GGFL01000134 MBW64312.1 CH477443 EAT40793.1 GGFJ01003665 MBW52806.1 EZ422979 ADD19255.1 GGFJ01003666 MBW52807.1 DS235813 EEB17458.1 GGFK01007878 MBW41199.1 JRES01000062 KNC34516.1 AXCM01000497 JQ042683 AEZ67828.1 JXJN01020367 AJWK01009480 PYGN01001609 PSN33756.1 JXUM01017367 JXUM01017368 KQ560481 KXJ82201.1 JXUM01088539 JXUM01088540 JXUM01088541 KQ563708 KXJ73459.1 CM000160 EDW98864.1 CM000364 EDX14310.1 CH902617 KPU80500.1 AE014297 AY061834 AAF56366.2 AAL27645.1 GANO01002094 JAB57777.1 CH954182 EDV53776.1 BT056260 ACL68707.1 GFDF01000939 JAV13145.1 BT003598 AAO39601.1 CM000070 EDY67871.2 CH940652 EDW59129.2 CH916377 EDV90665.1 ACPB03006555 QOIP01000011 RLU17264.1 GFTR01006954 JAW09472.1 GAHY01001763 JAA75747.1 GECL01001988 JAP04136.1 OUUW01000005 SPP80334.1 APCN01000594 CH479179 EDW25015.1 CVRI01000069 CRL06983.1 CH933806 EDW14751.2 CH964272 EDW84736.2 CP012526 ALC46780.1
KQ459582 KPI99044.1 AGBW02007651 OWR55079.1 KQ460651 KPJ13034.1 ODYU01008673 SOQ52455.1 KZ288204 PBC33322.1 GL887844 EGI69864.1 KQ978730 KYN28955.1 ADTU01022700 ADTU01022701 ADTU01022702 KQ982409 KYQ56676.1 KQ981986 KYN31240.1 KQ435784 KOX74552.1 GL766616 EFZ14930.1 PBC33320.1 GL443269 EFN62290.1 NEVH01016329 PNF25674.1 PNF25670.1 PNF25673.1 KQ435090 KZC14605.1 JR036511 JR036513 AEY57188.1 AEY57190.1 LBMM01003497 KMQ93464.1 KQ976745 KYM75427.1 KK853383 KDR07999.1 KQ414619 KOC67769.1 GAMC01004368 JAC02188.1 KK107105 EZA59527.1 GL451493 EFN79210.1 GDHF01030674 GDHF01002354 JAI21640.1 JAI49960.1 GAKP01007457 GAKP01007456 JAC51495.1 GEBQ01016594 JAT23383.1 GBXI01007433 JAD06859.1 NNAY01000414 OXU28577.1 GECZ01008465 JAS61304.1 AAAB01008859 EAA07616.5 UFQS01000993 UFQT01000993 SSX08310.1 SSX28333.1 AXCN02000138 ADMH02001019 ETN64436.1 GGFL01000133 MBW64311.1 GGFL01000134 MBW64312.1 CH477443 EAT40793.1 GGFJ01003665 MBW52806.1 EZ422979 ADD19255.1 GGFJ01003666 MBW52807.1 DS235813 EEB17458.1 GGFK01007878 MBW41199.1 JRES01000062 KNC34516.1 AXCM01000497 JQ042683 AEZ67828.1 JXJN01020367 AJWK01009480 PYGN01001609 PSN33756.1 JXUM01017367 JXUM01017368 KQ560481 KXJ82201.1 JXUM01088539 JXUM01088540 JXUM01088541 KQ563708 KXJ73459.1 CM000160 EDW98864.1 CM000364 EDX14310.1 CH902617 KPU80500.1 AE014297 AY061834 AAF56366.2 AAL27645.1 GANO01002094 JAB57777.1 CH954182 EDV53776.1 BT056260 ACL68707.1 GFDF01000939 JAV13145.1 BT003598 AAO39601.1 CM000070 EDY67871.2 CH940652 EDW59129.2 CH916377 EDV90665.1 ACPB03006555 QOIP01000011 RLU17264.1 GFTR01006954 JAW09472.1 GAHY01001763 JAA75747.1 GECL01001988 JAP04136.1 OUUW01000005 SPP80334.1 APCN01000594 CH479179 EDW25015.1 CVRI01000069 CRL06983.1 CH933806 EDW14751.2 CH964272 EDW84736.2 CP012526 ALC46780.1
Proteomes
UP000218220
UP000283053
UP000053268
UP000007151
UP000053240
UP000005203
+ More
UP000242457 UP000007755 UP000078492 UP000005205 UP000075809 UP000078541 UP000053105 UP000000311 UP000235965 UP000076502 UP000036403 UP000078540 UP000027135 UP000053825 UP000053097 UP000008237 UP000002358 UP000215335 UP000076407 UP000007062 UP000095300 UP000075886 UP000000673 UP000008820 UP000075903 UP000009046 UP000095301 UP000075902 UP000075882 UP000037069 UP000075883 UP000092460 UP000069272 UP000092443 UP000075920 UP000076408 UP000192221 UP000092461 UP000245037 UP000069940 UP000249989 UP000092445 UP000075881 UP000002282 UP000000304 UP000007801 UP000000803 UP000008711 UP000075884 UP000001819 UP000008792 UP000001070 UP000015103 UP000279307 UP000268350 UP000075840 UP000008744 UP000183832 UP000091820 UP000009192 UP000007798 UP000092553 UP000078200
UP000242457 UP000007755 UP000078492 UP000005205 UP000075809 UP000078541 UP000053105 UP000000311 UP000235965 UP000076502 UP000036403 UP000078540 UP000027135 UP000053825 UP000053097 UP000008237 UP000002358 UP000215335 UP000076407 UP000007062 UP000095300 UP000075886 UP000000673 UP000008820 UP000075903 UP000009046 UP000095301 UP000075902 UP000075882 UP000037069 UP000075883 UP000092460 UP000069272 UP000092443 UP000075920 UP000076408 UP000192221 UP000092461 UP000245037 UP000069940 UP000249989 UP000092445 UP000075881 UP000002282 UP000000304 UP000007801 UP000000803 UP000008711 UP000075884 UP000001819 UP000008792 UP000001070 UP000015103 UP000279307 UP000268350 UP000075840 UP000008744 UP000183832 UP000091820 UP000009192 UP000007798 UP000092553 UP000078200
Interpro
SUPFAM
SSF111331
SSF111331
Gene 3D
ProteinModelPortal
A0A2H1VAI5
A0A2A4JFF9
A0A3S2PD05
A0A194Q220
A0A212FMZ7
A0A0N0PC78
+ More
A0A2H1WHA7 A0A087ZP46 A0A2A3EPX6 F4W7J1 A0A195EL23 A0A158NPY5 A0A151X8H3 A0A195ESL5 A0A0N0U5B5 E9IWW3 A0A2A3ENP4 E2AWA0 A0A2J7QAS0 A0A2J7QAQ0 A0A2J7QAR0 A0A154PRQ6 V9I7U5 A0A0J7KT94 A0A195ATP6 A0A067QRZ5 A0A0L7RA97 W8BSR2 A0A026WU23 E2BYK5 A0A0K8U5I9 A0A034WBH5 A0A1B6LI78 A0A0A1X6D4 K7IW73 A0A232FE10 A0A1I8JW47 A0A1B6GFU4 Q7QD35 A0A336KT59 A0A1I8PFA1 A0A182Q707 W5JK02 A0A2M4CGM8 A0A2M4CG41 Q171Z3 A0A2M4BIC2 D3TNH7 A0A2M4BID8 A0A182UY32 E0VVQ2 A0A1I8M0S5 A0A2M4AK84 A0A182TQJ0 A0A182LG37 A0A0L0CQJ2 A0A182M739 H6WAR0 A0A1B0BTU3 A0A182FNT1 A0A1A9XDK3 A0A182VRR3 A0A182XV12 A0A1W4VKY4 A0A1B0CEZ4 A0A2P8XP31 A0A182GSS5 A0A1A9ZGD9 A0A182GU49 A0A182JXQ6 B4PV15 B4QU73 A0A0N8P1P5 Q9VC10 U5EV01 B3P6V6 B8A3W8 A0A1L8E3R1 A0A182NLH8 Q86NX1 B5DXD5 B4M653 B4JYA3 T1HSQ2 A0A3L8DAA6 A0A224XLJ6 R4FKN9 A0A0V0G8U0 A0A3B0JD79 A0A182HTZ1 B4G3K7 A0A1J1J3M9 A0A1A9WRR7 B4KBS4 B4NI44 A0A0M4EUJ5 A0A1A9VTW3
A0A2H1WHA7 A0A087ZP46 A0A2A3EPX6 F4W7J1 A0A195EL23 A0A158NPY5 A0A151X8H3 A0A195ESL5 A0A0N0U5B5 E9IWW3 A0A2A3ENP4 E2AWA0 A0A2J7QAS0 A0A2J7QAQ0 A0A2J7QAR0 A0A154PRQ6 V9I7U5 A0A0J7KT94 A0A195ATP6 A0A067QRZ5 A0A0L7RA97 W8BSR2 A0A026WU23 E2BYK5 A0A0K8U5I9 A0A034WBH5 A0A1B6LI78 A0A0A1X6D4 K7IW73 A0A232FE10 A0A1I8JW47 A0A1B6GFU4 Q7QD35 A0A336KT59 A0A1I8PFA1 A0A182Q707 W5JK02 A0A2M4CGM8 A0A2M4CG41 Q171Z3 A0A2M4BIC2 D3TNH7 A0A2M4BID8 A0A182UY32 E0VVQ2 A0A1I8M0S5 A0A2M4AK84 A0A182TQJ0 A0A182LG37 A0A0L0CQJ2 A0A182M739 H6WAR0 A0A1B0BTU3 A0A182FNT1 A0A1A9XDK3 A0A182VRR3 A0A182XV12 A0A1W4VKY4 A0A1B0CEZ4 A0A2P8XP31 A0A182GSS5 A0A1A9ZGD9 A0A182GU49 A0A182JXQ6 B4PV15 B4QU73 A0A0N8P1P5 Q9VC10 U5EV01 B3P6V6 B8A3W8 A0A1L8E3R1 A0A182NLH8 Q86NX1 B5DXD5 B4M653 B4JYA3 T1HSQ2 A0A3L8DAA6 A0A224XLJ6 R4FKN9 A0A0V0G8U0 A0A3B0JD79 A0A182HTZ1 B4G3K7 A0A1J1J3M9 A0A1A9WRR7 B4KBS4 B4NI44 A0A0M4EUJ5 A0A1A9VTW3
Ontologies
PATHWAY
GO
PANTHER
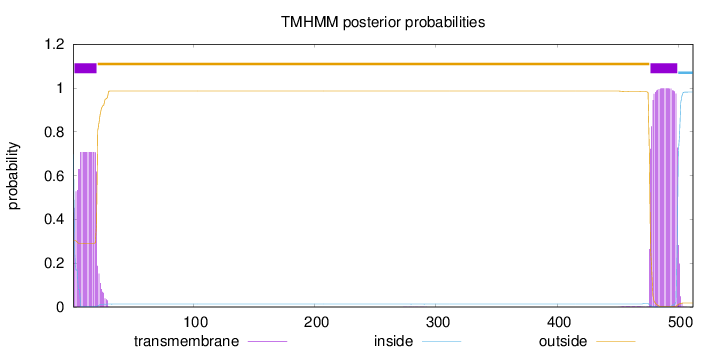
Topology
Length:
512
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
36.31497
Exp number, first 60 AAs:
13.36438
Total prob of N-in:
0.69734
POSSIBLE N-term signal
sequence
inside
1 - 1
TMhelix
2 - 20
outside
21 - 476
TMhelix
477 - 499
inside
500 - 512
Population Genetic Test Statistics
Pi
211.171097
Theta
159.952758
Tajima's D
1.504466
CLR
0.202128
CSRT
0.782310884455777
Interpretation
Uncertain
