Gene
KWMTBOMO09478
Pre Gene Modal
BGIBMGA012799
Annotation
PREDICTED:_uncharacterized_protein_LOC101743841_[Bombyx_mori]
Full name
Serine/threonine-protein phosphatase
Location in the cell
PlasmaMembrane Reliability : 3.166
Sequence
CDS
ATGAGTTTGAAAAGTACCCCGAATATGTCTAAGCGTGAGAAATTTCAACTAATTAACTCTGAAGTGCGTTCTTTTTACGAATATTGTAAAGAAGCGGGAATAAGTGACGAAGAAATGGATATTATATGTCGACCTCTGACGAATGCTGTGCGGAAAGCATACATAAGACGTTGGACAAGATTCTTTTTATGTATCATTATGGTATTGGCGTTGGGTTATACTGTGAGCCAAACGGATACATTCCAGTGGCACGCATCGGCTGCTGCAAGAATGCTGCTGATCAAGATCCTGCCCGTATGGGACTGGACACCACTATACTACAACAAGTGCTTGATAGAGAGATCTCAACCGCAGAGTTTAGATAGCGATAATGTTATAACGCCAATTGATTGCATTTCATGTGAGGCTGTCAAAAACGTTGCTCGCTTATCGGACACCAGTTACAGCGAAGTGTTCAATCATCATCTCATGCGTGGAGCCCCGGTCATCGTGGTAGATGCCTATTACGATTGGGCTAGCAGCGAGCTAAACCTCACCAGTTTTATAAGCGAAGACTCTAGACTGCGCGATTCCGTACCATGCAGGATCTTGACGAACATCAAAATTGCTCGCCAGCCCATGGACTTAGAGGAAATCATTTCCCGCATCACCAATACAGAGATTCCTAGTTGGTTCGTTCACTTCCAGAACTGCGATATCCACGCGGTAAAAACGTTTAGGATCCTCGCTCCGAGACCGTATTTCCTTTCCCCCCACATCCCCCCATCTCATTTCAACTGGCTGGTGCTGTCGAAAAACTATGATACTCACAGATACAAGTTTCTAGAGCTCGACGTGGGTCTAATCATAATGTCTCAATTAAAAGGAAAGTCCTTCGTTCAGTTGAGGCCTCGTGCACCCTGCGAGGAAGACTGTTACACCCTCAACGTAGAAGTGCAAGAGGGTGAAACTCTAGTCTTGGGTAATTCGCTGTGGGAATTTGGTTATTTACCAGGGAAGGGAGAGAATTTTGCGATCGTTACCGAAACGGATTGGGACTAG
Protein
MSLKSTPNMSKREKFQLINSEVRSFYEYCKEAGISDEEMDIICRPLTNAVRKAYIRRWTRFFLCIIMVLALGYTVSQTDTFQWHASAAARMLLIKILPVWDWTPLYYNKCLIERSQPQSLDSDNVITPIDCISCEAVKNVARLSDTSYSEVFNHHLMRGAPVIVVDAYYDWASSELNLTSFISEDSRLRDSVPCRILTNIKIARQPMDLEEIISRITNTEIPSWFVHFQNCDIHAVKTFRILAPRPYFLSPHIPPSHFNWLVLSKNYDTHRYKFLELDVGLIIMSQLKGKSFVQLRPRAPCEEDCYTLNVEVQEGETLVLGNSLWEFGYLPGKGENFAIVTETDWD
Summary
Catalytic Activity
H2O + O-phospho-L-seryl-[protein] = L-seryl-[protein] + phosphate
H2O + O-phospho-L-threonyl-[protein] = L-threonyl-[protein] + phosphate
H2O + O-phospho-L-threonyl-[protein] = L-threonyl-[protein] + phosphate
Similarity
Belongs to the PPP phosphatase family.
Belongs to the TRAFAC class TrmE-Era-EngA-EngB-Septin-like GTPase superfamily. Septin GTPase family.
Belongs to the TRAFAC class TrmE-Era-EngA-EngB-Septin-like GTPase superfamily. Septin GTPase family.
Uniprot
H9JTD7
A0A437BBW7
A0A2W1BS75
A0A2H1VBM9
A0A2A4JF54
A0A0N0PCH2
+ More
A0A0L7KSP9 A0A212FMW0 A0A194Q142 A0A067R8X5 A0A2J7PX40 A0A2A3ESG7 V9IC15 A0A088AUP2 A0A195DQZ2 F4X8B9 A0A195FDA7 A0A158NBS8 A0A151X9G8 E2ADJ5 A0A154PL08 E9IJI0 A0A1B6L4X3 A0A1B6EMH3 A0A026W2T0 A0A232ERL8 A0A151IMA7 E0VSP2 A0A195BGA1 A0A0C9QII6 A0A0N0BD63 A0A1B6CL03 A0A0P4VTE8 A0A1B6HS76 A0A023EY27 V9ICL6 A0A0L7QL18 A0A0A9ZET7 A0A0J7L269 A0A0V0GCA7 A0A2J7PX42 A0A2P8ZJL2 B4P4Z9 A0A2L2Y9H4 B4J8F9 A0A1I8PBE1 A0A1Y1JYX1 Q058Y4 A0A182P7E0 B4MPE9 B4HPN2 A0A182GGU0 A0A2T7NYX8 D2A4A7 A0A2P6KZN2 B4QDF7 R7TLG1 A0A226EI12 A0A084VZX6 A0A087U2L5 A0A182VY07 A0A1W4VRG5 B4KRS2 B3NRW5 B0W3K0 A0A423SRS2 A0A182MRB3 A0A336K1B6 A0A182T8H1 A0A182YP40 B3MG25 B5DZJ7 A0A3B0JU61 A0A182I8Z9 A0A182LAY4 Q7QG13 A0A3S1BS86 A0A182K5U1 A0A182UWR1 A0A182TE67 B4GIK8 A0A1W4WPT9 A0A0L0C704 B4MFN9 A0A0L8HT79 A0A0M4E9D0 A0A182N4J9 A0A1A9ZYS5 A0A182F952 T1J6H9 A0A3M6T909 A0A1B0ALJ9 A0A1A9YFX0 A0A1S3K2E9 A0A1A9UP04 A0A182R0T0 N6U8V2
A0A0L7KSP9 A0A212FMW0 A0A194Q142 A0A067R8X5 A0A2J7PX40 A0A2A3ESG7 V9IC15 A0A088AUP2 A0A195DQZ2 F4X8B9 A0A195FDA7 A0A158NBS8 A0A151X9G8 E2ADJ5 A0A154PL08 E9IJI0 A0A1B6L4X3 A0A1B6EMH3 A0A026W2T0 A0A232ERL8 A0A151IMA7 E0VSP2 A0A195BGA1 A0A0C9QII6 A0A0N0BD63 A0A1B6CL03 A0A0P4VTE8 A0A1B6HS76 A0A023EY27 V9ICL6 A0A0L7QL18 A0A0A9ZET7 A0A0J7L269 A0A0V0GCA7 A0A2J7PX42 A0A2P8ZJL2 B4P4Z9 A0A2L2Y9H4 B4J8F9 A0A1I8PBE1 A0A1Y1JYX1 Q058Y4 A0A182P7E0 B4MPE9 B4HPN2 A0A182GGU0 A0A2T7NYX8 D2A4A7 A0A2P6KZN2 B4QDF7 R7TLG1 A0A226EI12 A0A084VZX6 A0A087U2L5 A0A182VY07 A0A1W4VRG5 B4KRS2 B3NRW5 B0W3K0 A0A423SRS2 A0A182MRB3 A0A336K1B6 A0A182T8H1 A0A182YP40 B3MG25 B5DZJ7 A0A3B0JU61 A0A182I8Z9 A0A182LAY4 Q7QG13 A0A3S1BS86 A0A182K5U1 A0A182UWR1 A0A182TE67 B4GIK8 A0A1W4WPT9 A0A0L0C704 B4MFN9 A0A0L8HT79 A0A0M4E9D0 A0A182N4J9 A0A1A9ZYS5 A0A182F952 T1J6H9 A0A3M6T909 A0A1B0ALJ9 A0A1A9YFX0 A0A1S3K2E9 A0A1A9UP04 A0A182R0T0 N6U8V2
EC Number
3.1.3.16
Pubmed
19121390
28756777
26354079
26227816
22118469
24845553
+ More
21719571 21347285 20798317 21282665 24508170 30249741 28648823 20566863 27129103 25474469 25401762 29403074 17994087 17550304 26561354 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26483478 18362917 19820115 22936249 23254933 24438588 25244985 15632085 20966253 12364791 14747013 17210077 26108605 30382153 23537049
21719571 21347285 20798317 21282665 24508170 30249741 28648823 20566863 27129103 25474469 25401762 29403074 17994087 17550304 26561354 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26483478 18362917 19820115 22936249 23254933 24438588 25244985 15632085 20966253 12364791 14747013 17210077 26108605 30382153 23537049
EMBL
BABH01004081
RSAL01000093
RVE47890.1
KZ149935
PZC77181.1
ODYU01001508
+ More
SOQ37812.1 NWSH01001675 PCG70449.1 KQ460651 KPJ13035.1 JTDY01006335 KOB66091.1 AGBW02007651 OWR55081.1 KQ459582 KPI99043.1 KK852619 KDR20106.1 NEVH01020862 PNF20901.1 KZ288192 PBC34222.1 JR038277 AEY58192.1 KQ980581 KYN15266.1 GL888932 EGI57189.1 KQ981673 KYN38366.1 ADTU01011296 KQ982373 KYQ57012.1 GL438812 EFN68489.1 KQ434954 KZC12532.1 GL763802 EFZ19275.1 GEBQ01021272 JAT18705.1 GECZ01030642 JAS39127.1 KK107459 QOIP01000004 EZA50390.1 RLU23710.1 NNAY01002580 OXU21004.1 KQ977059 KYN06022.1 DS235755 EEB16398.1 KQ976500 KYM83185.1 GBYB01000407 JAG70174.1 KQ435883 KOX69698.1 GEDC01023253 JAS14045.1 GDKW01002373 JAI54222.1 GECU01030205 JAS77501.1 GBBI01004719 JAC13993.1 JR038276 AEY58191.1 KQ414934 KOC59264.1 GBHO01001173 JAG42431.1 LBMM01001173 KMQ96781.1 KMQ96786.1 GECL01000428 JAP05696.1 PNF20903.1 PYGN01000035 PSN56686.1 CM000158 EDW90720.1 IAAA01016478 LAA04793.1 CH916367 EDW02318.1 GEZM01099222 GEZM01099221 JAV53431.1 AE013599 BT029084 AAM68650.2 ABJ17017.1 CH963849 EDW73988.1 CH480816 EDW47616.1 JXUM01062338 KQ562195 KXJ76439.1 PZQS01000008 PVD26356.1 KQ971348 EFA05643.1 MWRG01003266 PRD31772.1 CM000362 CM002911 EDX06822.1 KMY93319.1 AMQN01012193 AMQN01012194 KB309360 ELT94683.1 LNIX01000004 OXA56226.1 ATLV01019028 KE525259 KFB43520.1 KK117863 KFM71604.1 CH933808 EDW08342.1 CH954179 EDV56267.1 DS231832 EDS31786.1 QCYY01002879 ROT66874.1 AXCM01006164 UFQS01000022 UFQT01000022 SSW97602.1 SSX17988.1 CH902619 EDV36720.1 CM000071 EDY69203.2 OUUW01000001 SPP74608.1 APCN01003142 AAAB01008844 EAA06103.5 RQTK01000036 RUS90255.1 CH479183 EDW36328.1 JRES01000930 KNC27214.1 CH940667 EDW57210.1 KQ417343 KOF92404.1 CP012524 ALC41496.1 JH431878 RCHS01004072 RMX37811.1 JXJN01031215 AXCN02000044 APGK01044194 APGK01044195 KB741021 ENN75027.1
SOQ37812.1 NWSH01001675 PCG70449.1 KQ460651 KPJ13035.1 JTDY01006335 KOB66091.1 AGBW02007651 OWR55081.1 KQ459582 KPI99043.1 KK852619 KDR20106.1 NEVH01020862 PNF20901.1 KZ288192 PBC34222.1 JR038277 AEY58192.1 KQ980581 KYN15266.1 GL888932 EGI57189.1 KQ981673 KYN38366.1 ADTU01011296 KQ982373 KYQ57012.1 GL438812 EFN68489.1 KQ434954 KZC12532.1 GL763802 EFZ19275.1 GEBQ01021272 JAT18705.1 GECZ01030642 JAS39127.1 KK107459 QOIP01000004 EZA50390.1 RLU23710.1 NNAY01002580 OXU21004.1 KQ977059 KYN06022.1 DS235755 EEB16398.1 KQ976500 KYM83185.1 GBYB01000407 JAG70174.1 KQ435883 KOX69698.1 GEDC01023253 JAS14045.1 GDKW01002373 JAI54222.1 GECU01030205 JAS77501.1 GBBI01004719 JAC13993.1 JR038276 AEY58191.1 KQ414934 KOC59264.1 GBHO01001173 JAG42431.1 LBMM01001173 KMQ96781.1 KMQ96786.1 GECL01000428 JAP05696.1 PNF20903.1 PYGN01000035 PSN56686.1 CM000158 EDW90720.1 IAAA01016478 LAA04793.1 CH916367 EDW02318.1 GEZM01099222 GEZM01099221 JAV53431.1 AE013599 BT029084 AAM68650.2 ABJ17017.1 CH963849 EDW73988.1 CH480816 EDW47616.1 JXUM01062338 KQ562195 KXJ76439.1 PZQS01000008 PVD26356.1 KQ971348 EFA05643.1 MWRG01003266 PRD31772.1 CM000362 CM002911 EDX06822.1 KMY93319.1 AMQN01012193 AMQN01012194 KB309360 ELT94683.1 LNIX01000004 OXA56226.1 ATLV01019028 KE525259 KFB43520.1 KK117863 KFM71604.1 CH933808 EDW08342.1 CH954179 EDV56267.1 DS231832 EDS31786.1 QCYY01002879 ROT66874.1 AXCM01006164 UFQS01000022 UFQT01000022 SSW97602.1 SSX17988.1 CH902619 EDV36720.1 CM000071 EDY69203.2 OUUW01000001 SPP74608.1 APCN01003142 AAAB01008844 EAA06103.5 RQTK01000036 RUS90255.1 CH479183 EDW36328.1 JRES01000930 KNC27214.1 CH940667 EDW57210.1 KQ417343 KOF92404.1 CP012524 ALC41496.1 JH431878 RCHS01004072 RMX37811.1 JXJN01031215 AXCN02000044 APGK01044194 APGK01044195 KB741021 ENN75027.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053240
UP000037510
UP000007151
+ More
UP000053268 UP000027135 UP000235965 UP000242457 UP000005203 UP000078492 UP000007755 UP000078541 UP000005205 UP000075809 UP000000311 UP000076502 UP000053097 UP000279307 UP000215335 UP000078542 UP000009046 UP000078540 UP000053105 UP000053825 UP000036403 UP000245037 UP000002282 UP000001070 UP000095300 UP000000803 UP000075885 UP000007798 UP000001292 UP000069940 UP000249989 UP000245119 UP000007266 UP000000304 UP000014760 UP000198287 UP000030765 UP000054359 UP000075920 UP000192221 UP000009192 UP000008711 UP000002320 UP000283509 UP000075883 UP000075901 UP000076408 UP000007801 UP000001819 UP000268350 UP000075840 UP000075882 UP000007062 UP000271974 UP000075881 UP000075903 UP000075902 UP000008744 UP000192223 UP000037069 UP000008792 UP000053454 UP000092553 UP000075884 UP000092445 UP000069272 UP000275408 UP000092460 UP000092443 UP000085678 UP000078200 UP000075886 UP000019118
UP000053268 UP000027135 UP000235965 UP000242457 UP000005203 UP000078492 UP000007755 UP000078541 UP000005205 UP000075809 UP000000311 UP000076502 UP000053097 UP000279307 UP000215335 UP000078542 UP000009046 UP000078540 UP000053105 UP000053825 UP000036403 UP000245037 UP000002282 UP000001070 UP000095300 UP000000803 UP000075885 UP000007798 UP000001292 UP000069940 UP000249989 UP000245119 UP000007266 UP000000304 UP000014760 UP000198287 UP000030765 UP000054359 UP000075920 UP000192221 UP000009192 UP000008711 UP000002320 UP000283509 UP000075883 UP000075901 UP000076408 UP000007801 UP000001819 UP000268350 UP000075840 UP000075882 UP000007062 UP000271974 UP000075881 UP000075903 UP000075902 UP000008744 UP000192223 UP000037069 UP000008792 UP000053454 UP000092553 UP000075884 UP000092445 UP000069272 UP000275408 UP000092460 UP000092443 UP000085678 UP000078200 UP000075886 UP000019118
Interpro
IPR006186
Ser/Thr-sp_prot-phosphatase
+ More
IPR037981 PPP1CC
IPR031675 STPPase_N
IPR004843 Calcineurin-like_PHP_ApaH
IPR029052 Metallo-depent_PP-like
IPR002833 PTH2
IPR023476 Pep_tRNA_hydro_II_dom_sf
IPR035979 RBD_domain_sf
IPR034353 ABT1/ESF2_RRM
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR016491 Septin
IPR030643 Septin4
IPR027417 P-loop_NTPase
IPR030379 G_SEPTIN_dom
IPR037981 PPP1CC
IPR031675 STPPase_N
IPR004843 Calcineurin-like_PHP_ApaH
IPR029052 Metallo-depent_PP-like
IPR002833 PTH2
IPR023476 Pep_tRNA_hydro_II_dom_sf
IPR035979 RBD_domain_sf
IPR034353 ABT1/ESF2_RRM
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR016491 Septin
IPR030643 Septin4
IPR027417 P-loop_NTPase
IPR030379 G_SEPTIN_dom
Gene 3D
ProteinModelPortal
H9JTD7
A0A437BBW7
A0A2W1BS75
A0A2H1VBM9
A0A2A4JF54
A0A0N0PCH2
+ More
A0A0L7KSP9 A0A212FMW0 A0A194Q142 A0A067R8X5 A0A2J7PX40 A0A2A3ESG7 V9IC15 A0A088AUP2 A0A195DQZ2 F4X8B9 A0A195FDA7 A0A158NBS8 A0A151X9G8 E2ADJ5 A0A154PL08 E9IJI0 A0A1B6L4X3 A0A1B6EMH3 A0A026W2T0 A0A232ERL8 A0A151IMA7 E0VSP2 A0A195BGA1 A0A0C9QII6 A0A0N0BD63 A0A1B6CL03 A0A0P4VTE8 A0A1B6HS76 A0A023EY27 V9ICL6 A0A0L7QL18 A0A0A9ZET7 A0A0J7L269 A0A0V0GCA7 A0A2J7PX42 A0A2P8ZJL2 B4P4Z9 A0A2L2Y9H4 B4J8F9 A0A1I8PBE1 A0A1Y1JYX1 Q058Y4 A0A182P7E0 B4MPE9 B4HPN2 A0A182GGU0 A0A2T7NYX8 D2A4A7 A0A2P6KZN2 B4QDF7 R7TLG1 A0A226EI12 A0A084VZX6 A0A087U2L5 A0A182VY07 A0A1W4VRG5 B4KRS2 B3NRW5 B0W3K0 A0A423SRS2 A0A182MRB3 A0A336K1B6 A0A182T8H1 A0A182YP40 B3MG25 B5DZJ7 A0A3B0JU61 A0A182I8Z9 A0A182LAY4 Q7QG13 A0A3S1BS86 A0A182K5U1 A0A182UWR1 A0A182TE67 B4GIK8 A0A1W4WPT9 A0A0L0C704 B4MFN9 A0A0L8HT79 A0A0M4E9D0 A0A182N4J9 A0A1A9ZYS5 A0A182F952 T1J6H9 A0A3M6T909 A0A1B0ALJ9 A0A1A9YFX0 A0A1S3K2E9 A0A1A9UP04 A0A182R0T0 N6U8V2
A0A0L7KSP9 A0A212FMW0 A0A194Q142 A0A067R8X5 A0A2J7PX40 A0A2A3ESG7 V9IC15 A0A088AUP2 A0A195DQZ2 F4X8B9 A0A195FDA7 A0A158NBS8 A0A151X9G8 E2ADJ5 A0A154PL08 E9IJI0 A0A1B6L4X3 A0A1B6EMH3 A0A026W2T0 A0A232ERL8 A0A151IMA7 E0VSP2 A0A195BGA1 A0A0C9QII6 A0A0N0BD63 A0A1B6CL03 A0A0P4VTE8 A0A1B6HS76 A0A023EY27 V9ICL6 A0A0L7QL18 A0A0A9ZET7 A0A0J7L269 A0A0V0GCA7 A0A2J7PX42 A0A2P8ZJL2 B4P4Z9 A0A2L2Y9H4 B4J8F9 A0A1I8PBE1 A0A1Y1JYX1 Q058Y4 A0A182P7E0 B4MPE9 B4HPN2 A0A182GGU0 A0A2T7NYX8 D2A4A7 A0A2P6KZN2 B4QDF7 R7TLG1 A0A226EI12 A0A084VZX6 A0A087U2L5 A0A182VY07 A0A1W4VRG5 B4KRS2 B3NRW5 B0W3K0 A0A423SRS2 A0A182MRB3 A0A336K1B6 A0A182T8H1 A0A182YP40 B3MG25 B5DZJ7 A0A3B0JU61 A0A182I8Z9 A0A182LAY4 Q7QG13 A0A3S1BS86 A0A182K5U1 A0A182UWR1 A0A182TE67 B4GIK8 A0A1W4WPT9 A0A0L0C704 B4MFN9 A0A0L8HT79 A0A0M4E9D0 A0A182N4J9 A0A1A9ZYS5 A0A182F952 T1J6H9 A0A3M6T909 A0A1B0ALJ9 A0A1A9YFX0 A0A1S3K2E9 A0A1A9UP04 A0A182R0T0 N6U8V2
Ontologies
PANTHER
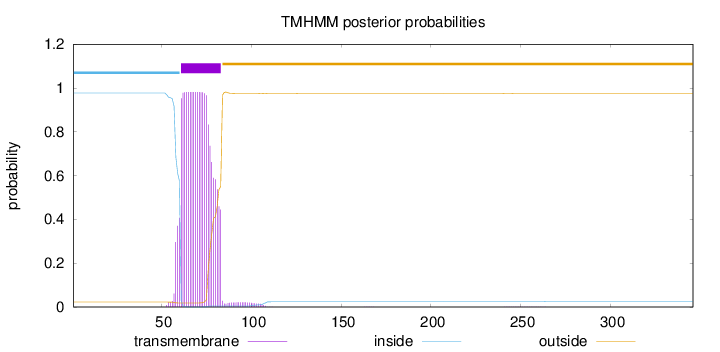
Topology
Length:
346
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.18286
Exp number, first 60 AAs:
1.21341
Total prob of N-in:
0.97687
inside
1 - 60
TMhelix
61 - 83
outside
84 - 346
Population Genetic Test Statistics
Pi
289.04602
Theta
219.928109
Tajima's D
1.127904
CLR
0
CSRT
0.692065396730164
Interpretation
Uncertain
