Pre Gene Modal
BGIBMGA012797
Annotation
PREDICTED:_vacuolar_protein_sorting-associated_protein_45_[Bombyx_mori]
Full name
Vacuolar protein sorting-associated protein 45
Location in the cell
Cytoplasmic Reliability : 1.348 Mitochondrial Reliability : 1.246 Nuclear Reliability : 1.536
Sequence
CDS
ATGAATGTGATACAAGCAGTTAAAATGTACATTACTAAAATGACGGAAGAGAGCGGCCCTGGAATGAAAGTGATATTAATGGATAAAGAAACTACCAGCATAGTGAGTATGGTGTACAGTCAATCGGAAATCTTACAGAAGGAAGTTTACTTATTCGAGAGGATAGACAGTCATGCCAAATGGGATAACATGAAACACATGAAATGCATAGTTTTTATACGTCCAACATCAGAGAACATAGCTCTGTTGTCACGGGAACTGAGAGATCCTAAATATGGTGTTTACTTTATATATTTCAGTAATGTAGTTTCTAAGGCAGACATCAAAACTTTGGCCGAATGTGATGAAGAGGAGGCTGTCAGAGAGGTCCAGGAAGTTTTTGCTGACTACCTGGCTGTGGACAGACACCTGTTCTCTTTCAATATTGTTGGATGCCTCCAAGGTCGGGTGTGGAACCAACAGCATCTCCAACGAGTTTCTCAAGGTCTGCTGGCGCTCCTCCTGTCTCTGAAGCGGCGAGCCTGCGTTCGCTACGAGGCAGCATCAGAACCCTGCGCTCGTCTCGCGGAGAGGGTCCGCGATCTGCTGAGGAGGGAGGCTGTGCTCATAGATAATAGCATCCCTTATAACGGAGAGCTGCCCCCTACTCAGCTGCTGATCATCGATAGGAGAAATGATCCCGTCACGCCCCTATTGAGTCAGTGGACCTACCAGGCGATGGTGCACGAGCTGCTGACGATAAACAACAACCGCGTGAGCCTGGCGCACGTGCCCGACGTGCCCAAGGACTTCAGGGAGGTGGTGCTCTCCTCGGAGCAGGACGACTTCTACGCGAAGAATCTTTACTCCAACTTCGGCGAGATCGGGCAAACCATCAAGTCTCTGATGGAGGAGTTTCAGACGAAAGCGAAGAGCAACCAGAAAGTGGAGAGCATCAGCGATATGAAAAATTTCGTGGAACAGTACCCACTCTTCAAGAAAATGTCTGGCACGGTGACGAAGCACGTGACCGTGGTGGGGGAGCTCTCGGCCTGTGTCGGCCGCCACAAGCTGCTCGAGGTCTCCGAGCTGGAGCAGGAGATCGTCTGCCAGTCAGACCACTCTAAACACCTGCAGCGGCTGAAGGCGCTGCTGGCGGACGGCGCGCTGCGGGACAGCGACGCGGCGCGGCTGGCGAGCCTGTACGCGCTGCGCTACGAGAAGCACGCGGGGCAGGGGGTGGGGGCGGCGCTGGAGCTGCTGCGGGCGCGGGGGGCGGCGGACGGGGCTCCGCTGGCCGCGCTCGAGTACGGGGGGGCGCACGCGCGACACGGGGACCTCTTCGGTCTGCAGGACGCGGTCAAGATCACCAACAGGCTCTTCAAGGGTCTGAACGGCGTCGAAAACATATACACGCAACACACGCCCCCCCTGAAGGACACGATCGAGGATCTGTTGAGGGGCAAGCTGCGTGAGGCCCAGTACCCCACGCTGGGCGGGGACGACGGCCGCCGCCCCACGCACGTCGTCGTGTTCGTGGTGGGCGGGGCCACGTACGAGGAGTCGCTGTGCGTGCACCAGATCAACAAGGCCAACCCGGGGGCCAAGGTGGTGCTCGGCGGGACCACCATACATAACTCGGCGTCATTCTTCGAAGAAATAAAGTCGGCTATGCAGGGCGTGCACAGGACGCACACCAAACATATTAGGAATGTTTAG
Protein
MNVIQAVKMYITKMTEESGPGMKVILMDKETTSIVSMVYSQSEILQKEVYLFERIDSHAKWDNMKHMKCIVFIRPTSENIALLSRELRDPKYGVYFIYFSNVVSKADIKTLAECDEEEAVREVQEVFADYLAVDRHLFSFNIVGCLQGRVWNQQHLQRVSQGLLALLLSLKRRACVRYEAASEPCARLAERVRDLLRREAVLIDNSIPYNGELPPTQLLIIDRRNDPVTPLLSQWTYQAMVHELLTINNNRVSLAHVPDVPKDFREVVLSSEQDDFYAKNLYSNFGEIGQTIKSLMEEFQTKAKSNQKVESISDMKNFVEQYPLFKKMSGTVTKHVTVVGELSACVGRHKLLEVSELEQEIVCQSDHSKHLQRLKALLADGALRDSDAARLASLYALRYEKHAGQGVGAALELLRARGAADGAPLAALEYGGAHARHGDLFGLQDAVKITNRLFKGLNGVENIYTQHTPPLKDTIEDLLRGKLREAQYPTLGGDDGRRPTHVVVFVVGGATYEESLCVHQINKANPGAKVVLGGTTIHNSASFFEEIKSAMQGVHRTHTKHIRNV
Summary
Subunit
Interacts with STX6 and ZFYVE20.
Interacts with ZFYVE20 (By similarity). Interacts with STX6.
Interacts with ZFYVE20 (By similarity). Interacts with STX6.
Similarity
Belongs to the STXBP/unc-18/SEC1 family.
Keywords
Complete proteome
Endosome
Golgi apparatus
Membrane
Phosphoprotein
Protein transport
Reference proteome
Transport
Feature
chain Vacuolar protein sorting-associated protein 45
Uniprot
A0A2H1V8P9
A0A437BBZ3
A0A194Q137
A0A2W1BU52
A0A212FMV9
A0A2A4JJS5
+ More
A0A067RVQ6 A0A2J7R528 A0A1B6FE66 A0A2A3ECD9 A0A088AER4 A0A151JQR5 F4WRE8 A0A195FIX5 A0A0C9RL68 A0A195BF99 A0A026W5U0 A0A2J7R520 A0A151IGB2 A0A232F3X6 A0A433T7Z4 A0A1S3GYE5 N6TUZ7 A0A0J7KBP7 A0A1B6CKF1 S4RHQ9 A0A1B0CL37 A0A158NUK8 E2AFV3 E9IRW2 A0A0L0C014 K7FKQ5 U4TYV2 Q9VHB5 B3M2G2 A0A452J4B8 B4PUQ8 Q295J1 A0A218UEV6 B4QWK4 A0A0K8V5C0 A0A3B0KCF9 A0A034VHX4 A0A1S3F3K4 A0A1U7R2D6 A0A250Y2Y1 A0A1I8ME64 O08700 A0A1W4VZM3 P97390 T1PJ43 Q0D2D9 Q3THX4 W8BJG9 B3NZQ1 A0A1A9W462 A0A087UGW9 H3AZF0 D3TNU1 A0A1A9Z521 B4NB39 Q5XHB0 A0A1U7SKM3 A0A293LGR7 G3TKR4 D7EHQ8 A0A2Y9E9K1 G3VWS6 A0A3Q2HGR5 M3WCQ2 A0A1S3A9R4 F6W1A0 U3BRW1 F7AE31 A0A1D5R9F9 F1SDG5 U6CVG7 A0A2Y9L6K3 G9KXF7 M3Y200 F7ILP1 U3EPE1 A0A2I2Y2Z4 A0A2K5EN41 A0A2Y9GWN4 A0A3Q7QXK1 A0A383ZSF3 G3H2U9 G3SCA3 A0A452E4J0 A0A340XW22 A0A2K6U2P4 V4AN63 A0A3P8SYD4 L8IKS5 A0A3Q7N361 A0A2R9BUS7 A0A2R9BVC6 A0A096N7B5 A0A2Y9EX89
A0A067RVQ6 A0A2J7R528 A0A1B6FE66 A0A2A3ECD9 A0A088AER4 A0A151JQR5 F4WRE8 A0A195FIX5 A0A0C9RL68 A0A195BF99 A0A026W5U0 A0A2J7R520 A0A151IGB2 A0A232F3X6 A0A433T7Z4 A0A1S3GYE5 N6TUZ7 A0A0J7KBP7 A0A1B6CKF1 S4RHQ9 A0A1B0CL37 A0A158NUK8 E2AFV3 E9IRW2 A0A0L0C014 K7FKQ5 U4TYV2 Q9VHB5 B3M2G2 A0A452J4B8 B4PUQ8 Q295J1 A0A218UEV6 B4QWK4 A0A0K8V5C0 A0A3B0KCF9 A0A034VHX4 A0A1S3F3K4 A0A1U7R2D6 A0A250Y2Y1 A0A1I8ME64 O08700 A0A1W4VZM3 P97390 T1PJ43 Q0D2D9 Q3THX4 W8BJG9 B3NZQ1 A0A1A9W462 A0A087UGW9 H3AZF0 D3TNU1 A0A1A9Z521 B4NB39 Q5XHB0 A0A1U7SKM3 A0A293LGR7 G3TKR4 D7EHQ8 A0A2Y9E9K1 G3VWS6 A0A3Q2HGR5 M3WCQ2 A0A1S3A9R4 F6W1A0 U3BRW1 F7AE31 A0A1D5R9F9 F1SDG5 U6CVG7 A0A2Y9L6K3 G9KXF7 M3Y200 F7ILP1 U3EPE1 A0A2I2Y2Z4 A0A2K5EN41 A0A2Y9GWN4 A0A3Q7QXK1 A0A383ZSF3 G3H2U9 G3SCA3 A0A452E4J0 A0A340XW22 A0A2K6U2P4 V4AN63 A0A3P8SYD4 L8IKS5 A0A3Q7N361 A0A2R9BUS7 A0A2R9BVC6 A0A096N7B5 A0A2Y9EX89
Pubmed
26354079
28756777
22118469
24845553
21719571
24508170
+ More
30249741 28648823 23537049 21347285 20798317 21282665 26108605 17381049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 28562605 17550304 15632085 25348373 28087693 25315136 9106478 15489334 22673903 9045632 21183079 10349636 11042159 11076861 11217851 12466851 16141073 24495485 9215903 20353571 27762356 18362917 19820115 21709235 19892987 17975172 25243066 17495919 17431167 23236062 22398555 21804562 23254933 22751099 22722832
30249741 28648823 23537049 21347285 20798317 21282665 26108605 17381049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 28562605 17550304 15632085 25348373 28087693 25315136 9106478 15489334 22673903 9045632 21183079 10349636 11042159 11076861 11217851 12466851 16141073 24495485 9215903 20353571 27762356 18362917 19820115 21709235 19892987 17975172 25243066 17495919 17431167 23236062 22398555 21804562 23254933 22751099 22722832
EMBL
ODYU01001242
SOQ37187.1
RSAL01000093
RVE47892.1
KQ459582
KPI99038.1
+ More
KZ149935 PZC77185.1 AGBW02007651 OWR55084.1 NWSH01001250 PCG71978.1 KK852433 KDR23959.1 NEVH01007395 PNF35935.1 GECZ01021253 JAS48516.1 KZ288301 PBC28846.1 KQ978660 KYN29396.1 GL888285 EGI63219.1 KQ981522 KYN40615.1 GBYB01007699 GBYB01008985 GBYB01008987 JAG77466.1 JAG78752.1 JAG78754.1 KQ976504 KYM82867.1 KK107390 QOIP01000007 EZA51435.1 RLU20929.1 PNF35933.1 KQ977771 KYN00033.1 NNAY01001090 OXU25170.1 RQTK01000564 RUS77638.1 APGK01053605 KB741231 ENN72221.1 LBMM01010090 KMQ87679.1 GEDC01027041 GEDC01023406 JAS10257.1 JAS13892.1 AJWK01016879 ADTU01026591 ADTU01026592 GL439158 EFN67702.1 GL765283 EFZ16590.1 JRES01001097 KNC25576.1 AGCU01163926 AGCU01163927 AGCU01163928 KB631770 ERL86007.1 AE014297 AY119186 AAF54403.1 AAM51046.1 CH902617 EDV43415.1 CM000160 EDW96675.1 CM000070 EAL28720.1 MUZQ01000382 OWK52000.1 CM000364 EDX13608.1 GDHF01018549 JAI33765.1 OUUW01000008 SPP83879.1 GAKP01016848 JAC42104.1 GFFV01001974 JAV37971.1 U81160 BC081705 U66865 BC012691 BC058528 KA648704 AFP63333.1 BC121938 AAI21939.1 AK146190 AK168101 BAE26966.1 BAE40072.1 GAMC01007663 JAB98892.1 CH954181 EDV49761.1 KK119738 KFM76608.1 AFYH01122252 AFYH01122253 AFYH01122254 AFYH01122255 AFYH01122256 CCAG010000864 CCAG010006733 EZ423093 ADD19369.1 CH964232 EDW81003.1 BC084162 CM004481 AAH84162.1 OCT66501.1 GFWV01002338 MAA27068.1 KQ973114 EFA12148.2 AEFK01176820 AEFK01176821 AEFK01176822 AEFK01176823 AANG04001280 GAMT01007483 GAMS01007423 GAMR01010755 GAMQ01001506 JAB04378.1 JAB15713.1 JAB23177.1 JAB40345.1 JSUE03002352 JSUE03002353 AEMK02000022 HAAF01002600 CCP74426.1 JP020988 AES09586.1 AEYP01089787 GAMP01008601 JAB44154.1 CABD030006308 CABD030006309 CABD030006310 CABD030006311 CABD030006312 JH000120 EGV94786.1 LWLT01000004 KB199650 ESP05609.1 JH881048 ELR56931.1 AJFE02058615 AJFE02058616 AJFE02058617 AJFE02058618 AJFE02058619 AJFE02058620 AJFE02058621 AJFE02058622 AHZZ02016811
KZ149935 PZC77185.1 AGBW02007651 OWR55084.1 NWSH01001250 PCG71978.1 KK852433 KDR23959.1 NEVH01007395 PNF35935.1 GECZ01021253 JAS48516.1 KZ288301 PBC28846.1 KQ978660 KYN29396.1 GL888285 EGI63219.1 KQ981522 KYN40615.1 GBYB01007699 GBYB01008985 GBYB01008987 JAG77466.1 JAG78752.1 JAG78754.1 KQ976504 KYM82867.1 KK107390 QOIP01000007 EZA51435.1 RLU20929.1 PNF35933.1 KQ977771 KYN00033.1 NNAY01001090 OXU25170.1 RQTK01000564 RUS77638.1 APGK01053605 KB741231 ENN72221.1 LBMM01010090 KMQ87679.1 GEDC01027041 GEDC01023406 JAS10257.1 JAS13892.1 AJWK01016879 ADTU01026591 ADTU01026592 GL439158 EFN67702.1 GL765283 EFZ16590.1 JRES01001097 KNC25576.1 AGCU01163926 AGCU01163927 AGCU01163928 KB631770 ERL86007.1 AE014297 AY119186 AAF54403.1 AAM51046.1 CH902617 EDV43415.1 CM000160 EDW96675.1 CM000070 EAL28720.1 MUZQ01000382 OWK52000.1 CM000364 EDX13608.1 GDHF01018549 JAI33765.1 OUUW01000008 SPP83879.1 GAKP01016848 JAC42104.1 GFFV01001974 JAV37971.1 U81160 BC081705 U66865 BC012691 BC058528 KA648704 AFP63333.1 BC121938 AAI21939.1 AK146190 AK168101 BAE26966.1 BAE40072.1 GAMC01007663 JAB98892.1 CH954181 EDV49761.1 KK119738 KFM76608.1 AFYH01122252 AFYH01122253 AFYH01122254 AFYH01122255 AFYH01122256 CCAG010000864 CCAG010006733 EZ423093 ADD19369.1 CH964232 EDW81003.1 BC084162 CM004481 AAH84162.1 OCT66501.1 GFWV01002338 MAA27068.1 KQ973114 EFA12148.2 AEFK01176820 AEFK01176821 AEFK01176822 AEFK01176823 AANG04001280 GAMT01007483 GAMS01007423 GAMR01010755 GAMQ01001506 JAB04378.1 JAB15713.1 JAB23177.1 JAB40345.1 JSUE03002352 JSUE03002353 AEMK02000022 HAAF01002600 CCP74426.1 JP020988 AES09586.1 AEYP01089787 GAMP01008601 JAB44154.1 CABD030006308 CABD030006309 CABD030006310 CABD030006311 CABD030006312 JH000120 EGV94786.1 LWLT01000004 KB199650 ESP05609.1 JH881048 ELR56931.1 AJFE02058615 AJFE02058616 AJFE02058617 AJFE02058618 AJFE02058619 AJFE02058620 AJFE02058621 AJFE02058622 AHZZ02016811
Proteomes
UP000283053
UP000053268
UP000007151
UP000218220
UP000027135
UP000235965
+ More
UP000242457 UP000005203 UP000078492 UP000007755 UP000078541 UP000078540 UP000053097 UP000279307 UP000078542 UP000215335 UP000271974 UP000085678 UP000019118 UP000036403 UP000245300 UP000092461 UP000005205 UP000000311 UP000037069 UP000007267 UP000030742 UP000000803 UP000007801 UP000291020 UP000002282 UP000001819 UP000197619 UP000000304 UP000268350 UP000081671 UP000189706 UP000095301 UP000002494 UP000192221 UP000000589 UP000008711 UP000091820 UP000054359 UP000008672 UP000092444 UP000092445 UP000007798 UP000186698 UP000189705 UP000007646 UP000007266 UP000248480 UP000007648 UP000002281 UP000011712 UP000079721 UP000008225 UP000002280 UP000006718 UP000008227 UP000248482 UP000000715 UP000001519 UP000233020 UP000248481 UP000286640 UP000261681 UP000001075 UP000291000 UP000265300 UP000233220 UP000030746 UP000265080 UP000286641 UP000240080 UP000028761 UP000248484
UP000242457 UP000005203 UP000078492 UP000007755 UP000078541 UP000078540 UP000053097 UP000279307 UP000078542 UP000215335 UP000271974 UP000085678 UP000019118 UP000036403 UP000245300 UP000092461 UP000005205 UP000000311 UP000037069 UP000007267 UP000030742 UP000000803 UP000007801 UP000291020 UP000002282 UP000001819 UP000197619 UP000000304 UP000268350 UP000081671 UP000189706 UP000095301 UP000002494 UP000192221 UP000000589 UP000008711 UP000091820 UP000054359 UP000008672 UP000092444 UP000092445 UP000007798 UP000186698 UP000189705 UP000007646 UP000007266 UP000248480 UP000007648 UP000002281 UP000011712 UP000079721 UP000008225 UP000002280 UP000006718 UP000008227 UP000248482 UP000000715 UP000001519 UP000233020 UP000248481 UP000286640 UP000261681 UP000001075 UP000291000 UP000265300 UP000233220 UP000030746 UP000265080 UP000286641 UP000240080 UP000028761 UP000248484
Pfam
PF00995 Sec1
SUPFAM
SSF56815
SSF56815
Gene 3D
ProteinModelPortal
A0A2H1V8P9
A0A437BBZ3
A0A194Q137
A0A2W1BU52
A0A212FMV9
A0A2A4JJS5
+ More
A0A067RVQ6 A0A2J7R528 A0A1B6FE66 A0A2A3ECD9 A0A088AER4 A0A151JQR5 F4WRE8 A0A195FIX5 A0A0C9RL68 A0A195BF99 A0A026W5U0 A0A2J7R520 A0A151IGB2 A0A232F3X6 A0A433T7Z4 A0A1S3GYE5 N6TUZ7 A0A0J7KBP7 A0A1B6CKF1 S4RHQ9 A0A1B0CL37 A0A158NUK8 E2AFV3 E9IRW2 A0A0L0C014 K7FKQ5 U4TYV2 Q9VHB5 B3M2G2 A0A452J4B8 B4PUQ8 Q295J1 A0A218UEV6 B4QWK4 A0A0K8V5C0 A0A3B0KCF9 A0A034VHX4 A0A1S3F3K4 A0A1U7R2D6 A0A250Y2Y1 A0A1I8ME64 O08700 A0A1W4VZM3 P97390 T1PJ43 Q0D2D9 Q3THX4 W8BJG9 B3NZQ1 A0A1A9W462 A0A087UGW9 H3AZF0 D3TNU1 A0A1A9Z521 B4NB39 Q5XHB0 A0A1U7SKM3 A0A293LGR7 G3TKR4 D7EHQ8 A0A2Y9E9K1 G3VWS6 A0A3Q2HGR5 M3WCQ2 A0A1S3A9R4 F6W1A0 U3BRW1 F7AE31 A0A1D5R9F9 F1SDG5 U6CVG7 A0A2Y9L6K3 G9KXF7 M3Y200 F7ILP1 U3EPE1 A0A2I2Y2Z4 A0A2K5EN41 A0A2Y9GWN4 A0A3Q7QXK1 A0A383ZSF3 G3H2U9 G3SCA3 A0A452E4J0 A0A340XW22 A0A2K6U2P4 V4AN63 A0A3P8SYD4 L8IKS5 A0A3Q7N361 A0A2R9BUS7 A0A2R9BVC6 A0A096N7B5 A0A2Y9EX89
A0A067RVQ6 A0A2J7R528 A0A1B6FE66 A0A2A3ECD9 A0A088AER4 A0A151JQR5 F4WRE8 A0A195FIX5 A0A0C9RL68 A0A195BF99 A0A026W5U0 A0A2J7R520 A0A151IGB2 A0A232F3X6 A0A433T7Z4 A0A1S3GYE5 N6TUZ7 A0A0J7KBP7 A0A1B6CKF1 S4RHQ9 A0A1B0CL37 A0A158NUK8 E2AFV3 E9IRW2 A0A0L0C014 K7FKQ5 U4TYV2 Q9VHB5 B3M2G2 A0A452J4B8 B4PUQ8 Q295J1 A0A218UEV6 B4QWK4 A0A0K8V5C0 A0A3B0KCF9 A0A034VHX4 A0A1S3F3K4 A0A1U7R2D6 A0A250Y2Y1 A0A1I8ME64 O08700 A0A1W4VZM3 P97390 T1PJ43 Q0D2D9 Q3THX4 W8BJG9 B3NZQ1 A0A1A9W462 A0A087UGW9 H3AZF0 D3TNU1 A0A1A9Z521 B4NB39 Q5XHB0 A0A1U7SKM3 A0A293LGR7 G3TKR4 D7EHQ8 A0A2Y9E9K1 G3VWS6 A0A3Q2HGR5 M3WCQ2 A0A1S3A9R4 F6W1A0 U3BRW1 F7AE31 A0A1D5R9F9 F1SDG5 U6CVG7 A0A2Y9L6K3 G9KXF7 M3Y200 F7ILP1 U3EPE1 A0A2I2Y2Z4 A0A2K5EN41 A0A2Y9GWN4 A0A3Q7QXK1 A0A383ZSF3 G3H2U9 G3SCA3 A0A452E4J0 A0A340XW22 A0A2K6U2P4 V4AN63 A0A3P8SYD4 L8IKS5 A0A3Q7N361 A0A2R9BUS7 A0A2R9BVC6 A0A096N7B5 A0A2Y9EX89
PDB
4CCA
E-value=2.68387e-17,
Score=219
Ontologies
GO
PANTHER
Topology
Subcellular location
Golgi apparatus membrane
Associated with Golgi/endosomal vesicles and the trans-Golgi network. With evidence from 2 publications.
Endosome membrane Associated with Golgi/endosomal vesicles and the trans-Golgi network. With evidence from 2 publications.
Endosome membrane Associated with Golgi/endosomal vesicles and the trans-Golgi network. With evidence from 2 publications.
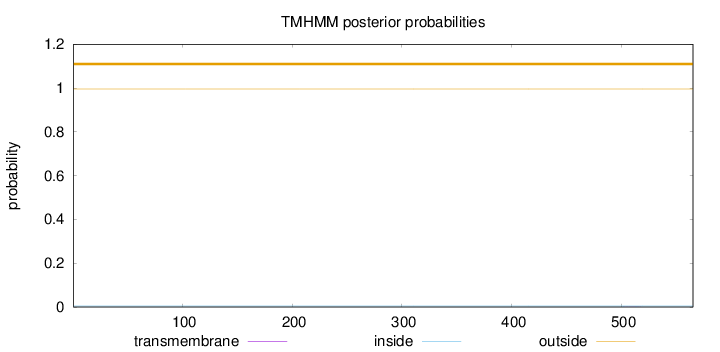
Length:
565
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00617
Exp number, first 60 AAs:
0.00056
Total prob of N-in:
0.00441
outside
1 - 565
Population Genetic Test Statistics
Pi
18.723674
Theta
20.908079
Tajima's D
-1.308068
CLR
2021.112215
CSRT
0.0839958002099895
Interpretation
Possibly Positive selection
