Gene
KWMTBOMO09474 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013063
Annotation
PREDICTED:_malate_dehydrogenase?_mitochondrial_[Amyelois_transitella]
Full name
Malate dehydrogenase
Location in the cell
Mitochondrial Reliability : 3.115
Sequence
CDS
ATGTTCTCCCGCGCCCTAAAACCTGCCGCCCTTGCTGTACAAAATGGTGCAAAGAACTTTTCCACCACATCACAGAGGAACTTTAAAGTGGTGGTCGCTGGTGCCGCCGGTGGTATCGGCCAGCCTTTGGCCCTTCTACTGAAGCAGAATCCTCTGGTGACCAGGCTGGCTTTATACGACATAGCGCCTGTGACCCCCGGCGTCGCAGCCGACCTTTCCCACATGAACACCCCAGCCAAGGTCAGCGGCCACAAGGGACCTGAGGAGCTATCAGCAGCCATCAAAGATGCTGATGTTGTAGTCATCCCAGCAGGAGTACCCCGTAAACCGGGAATGACCCGTGATGACCTATTCAACACTAATGCTTCCATTGTCCGTGACATTGCTTTGTCCATTGCTCAGAATGCACCCAAAGCTATTGTGGCCATCATCACAAACCCTGTTAACTCTACAGTGCCTATTGCCTCTGAAGTGCTCAAGAAGGCGGGAGTGTACGACCCGAACCGCGTGCTCGGCGTCACCACTCTGGACGTAGTCCGCGCTGCCACCTTCGTCGGCGAGATCAATGGCGTGGACCCGACCTCCGTCGCCGTCCCCGTCATCGGCGGTCACTCCGGCATCACCATCATCCCCATCCTGTCCCAGTGCCAGCCGGCCCTCAAGCTGAGTGACCCGGCCAAGATCAAGGCGCTCACCGAAAGGATCCAGGAGGCTGGTACCGAAGTTGTGAAGGCGAAGGCCGGCGGCGGCTCCGCGACCCTCTCCATGGCGTACGCCGGCGCGCGGCTCACGGCCTCCGTGCTCAGGGGACTGAAGGGCGAGTCCAACGTGGTGGAGTGCGCCTACGTCAAATCGAGCCTCACGGAGGCCACGTACTTCGCCAGCCCCGTGGTGCTCGGCAAGAGCGGCGTCGAGAAGGTCCTCGGCTACGGGACCCTCAACGAATTCGAGCAGCAACTCCTGAAGGCGGCCCTCCCTGAACTCGCCAAGAACATCAAGACCGGCGAGGACTTCGCCAAGAAGTAA
Protein
MFSRALKPAALAVQNGAKNFSTTSQRNFKVVVAGAAGGIGQPLALLLKQNPLVTRLALYDIAPVTPGVAADLSHMNTPAKVSGHKGPEELSAAIKDADVVVIPAGVPRKPGMTRDDLFNTNASIVRDIALSIAQNAPKAIVAIITNPVNSTVPIASEVLKKAGVYDPNRVLGVTTLDVVRAATFVGEINGVDPTSVAVPVIGGHSGITIIPILSQCQPALKLSDPAKIKALTERIQEAGTEVVKAKAGGGSATLSMAYAGARLTASVLRGLKGESNVVECAYVKSSLTEATYFASPVVLGKSGVEKVLGYGTLNEFEQQLLKAALPELAKNIKTGEDFAKK
Summary
Catalytic Activity
(S)-malate + NAD(+) = H(+) + NADH + oxaloacetate
Similarity
Belongs to the LDH/MDH superfamily.
Uniprot
H9JU51
A0A3S2NCY2
A0A212FMW7
A0A0N1IFM5
A0A194Q0C9
A0A2A4K4Z0
+ More
A0A2W1BWY1 I4DQC2 A0A2H1V8P8 A0A232F4J8 K7J833 A0A2P8YT87 A0A411G6K4 E2B525 E9IT62 A0A1L8EI00 A0A195E5P6 E2ADN0 A0A195C8W5 A0A0J7NNS3 A0A2Z5RE49 A0A2J7Q6U6 F4WPA4 A0A1D1XXI6 Q4PP90 A0A0L0BXC1 A0A336KS61 A0A1Y1KFL2 D2A663 A0A026WP45 A0A154PN63 A0A2P0XJ03 U5EL58 A0A182JME3 A0A0K8TPD6 A0A158NR67 A0A182PC32 A0A084VRY1 N6TTQ3 A0A1I9WLG5 J3JXT9 A0A182NZ02 A0A182MFG3 A0A182R1J5 A0A182QV09 A0A182VQY7 A0A0P6AFG0 A0A1B6JRY2 A0A1B6F1E5 A0A182S6J7 A0A2Y9D3K4 A0A182UXV3 A0A182U2M5 A0A182KM35 Q7PYE7 A0A182IIP6 A0A0P5EE84 A0A0A1XDJ4 A0A1B6LGI7 A0A151WME8 A0A182JQR4 A0A0P5L8R4 T1DMZ1 A0A2M4A2E0 A0A2M4BTF4 W5J4N2 A0A182FQJ7 A0A2M4CT91 W8BVV6 A0A2M4BT81 A0A0P6DCQ6 V5SJ21 A0A1L8EEQ2 A0A2M3YZS5 A0A1J1IGX3 A0A2M3YZM4 E9HGX0 A0A182GNQ1 A0A1L8EEK1 A0A1I8PUI8 T1PH40 A0A0K8VWB8 A0A0C9QFA6 A0A034VDU0 A0A023EQ02 B4JI52 A0A1S4FIY1 A0A1A9YNS9 A0A131XGD2 B3MTR2 Q293U0 B4GLM6 B4M0W4 A0A310SPT5 B4K7H4 A0A1B0BFC4 A0A3B0KCD1 B0WMC8 A0A0M4EPV2 Q16ZI5
A0A2W1BWY1 I4DQC2 A0A2H1V8P8 A0A232F4J8 K7J833 A0A2P8YT87 A0A411G6K4 E2B525 E9IT62 A0A1L8EI00 A0A195E5P6 E2ADN0 A0A195C8W5 A0A0J7NNS3 A0A2Z5RE49 A0A2J7Q6U6 F4WPA4 A0A1D1XXI6 Q4PP90 A0A0L0BXC1 A0A336KS61 A0A1Y1KFL2 D2A663 A0A026WP45 A0A154PN63 A0A2P0XJ03 U5EL58 A0A182JME3 A0A0K8TPD6 A0A158NR67 A0A182PC32 A0A084VRY1 N6TTQ3 A0A1I9WLG5 J3JXT9 A0A182NZ02 A0A182MFG3 A0A182R1J5 A0A182QV09 A0A182VQY7 A0A0P6AFG0 A0A1B6JRY2 A0A1B6F1E5 A0A182S6J7 A0A2Y9D3K4 A0A182UXV3 A0A182U2M5 A0A182KM35 Q7PYE7 A0A182IIP6 A0A0P5EE84 A0A0A1XDJ4 A0A1B6LGI7 A0A151WME8 A0A182JQR4 A0A0P5L8R4 T1DMZ1 A0A2M4A2E0 A0A2M4BTF4 W5J4N2 A0A182FQJ7 A0A2M4CT91 W8BVV6 A0A2M4BT81 A0A0P6DCQ6 V5SJ21 A0A1L8EEQ2 A0A2M3YZS5 A0A1J1IGX3 A0A2M3YZM4 E9HGX0 A0A182GNQ1 A0A1L8EEK1 A0A1I8PUI8 T1PH40 A0A0K8VWB8 A0A0C9QFA6 A0A034VDU0 A0A023EQ02 B4JI52 A0A1S4FIY1 A0A1A9YNS9 A0A131XGD2 B3MTR2 Q293U0 B4GLM6 B4M0W4 A0A310SPT5 B4K7H4 A0A1B0BFC4 A0A3B0KCD1 B0WMC8 A0A0M4EPV2 Q16ZI5
EC Number
1.1.1.37
Pubmed
19121390
22118469
26354079
28756777
22651552
28648823
+ More
20075255 29403074 20798317 21282665 26760975 21719571 26108605 28004739 18362917 19820115 24508170 26369729 21347285 24438588 23537049 27538518 22516182 20966253 12364791 14747013 17210077 25830018 20920257 23761445 24495485 21292972 26483478 25315136 25348373 24945155 17994087 28049606 15632085 17510324
20075255 29403074 20798317 21282665 26760975 21719571 26108605 28004739 18362917 19820115 24508170 26369729 21347285 24438588 23537049 27538518 22516182 20966253 12364791 14747013 17210077 25830018 20920257 23761445 24495485 21292972 26483478 25315136 25348373 24945155 17994087 28049606 15632085 17510324
EMBL
BABH01004084
BABH01004085
RSAL01000093
RVE47895.1
AGBW02007651
OWR55085.1
+ More
KQ460651 KPJ13039.1 KQ459582 KPI99037.1 NWSH01000176 PCG78732.1 KZ149935 PZC77186.1 AK404038 BAM20112.1 ODYU01001242 SOQ37186.1 NNAY01000986 OXU25605.1 PYGN01000374 PSN47461.1 MH365706 QBB01515.1 GL445685 EFN89233.1 GL765434 EFZ16256.1 GFDG01000570 JAV18229.1 KQ979608 KYN20488.1 GL438820 EFN68392.1 KQ978081 KYM97140.1 LBMM01002952 KMQ94145.1 FX983743 BAX07242.1 NEVH01017450 PNF24298.1 GL888243 EGI64085.1 GDJX01020827 JAT47109.1 DQ062223 AAY63978.1 JRES01001197 KNC24655.1 UFQS01000662 UFQT01000490 UFQT01000662 SSX05893.1 SSX24725.1 GEZM01087903 JAV58405.1 KQ971346 EFA04972.1 KK107139 EZA57723.1 KQ434960 KZC12660.1 KY661311 AVA17398.1 GANO01001560 JAB58311.1 GDAI01001597 JAI16006.1 ADTU01023767 ATLV01015785 KE525036 KFB40725.1 APGK01019088 KB740092 KB632429 ENN81433.1 ERL95462.1 KU932349 APA33985.1 BT128060 AEE63021.1 AXCM01001692 AXCN02000191 GDIQ01191327 GDIP01043456 GDIQ01037520 LRGB01000626 JAK60398.1 JAM60259.1 KZS17437.1 GECU01005761 JAT01946.1 GECZ01025697 JAS44072.1 AAAB01008987 EAA01572.4 APCN01002072 GDIQ01270961 JAJ80763.1 GBXI01004828 JAD09464.1 GEBQ01017166 JAT22811.1 KQ982944 KYQ49026.1 GDIQ01178465 JAK73260.1 GAMD01003320 JAA98270.1 GGFK01001594 MBW34915.1 GGFJ01007143 MBW56284.1 ADMH02002134 GGFL01004373 ETN58378.1 MBW68551.1 GGFL01004365 MBW68543.1 GAMC01003193 JAC03363.1 GGFJ01007144 MBW56285.1 GDIQ01082614 JAN12123.1 KF647639 AHB50501.1 GFDG01001643 JAV17156.1 GGFM01000967 MBW21718.1 CVRI01000051 CRK99511.1 GGFM01000971 MBW21722.1 GL732643 EFX69032.1 JXUM01076841 JXUM01076842 KQ562972 KXJ74736.1 GFDG01001652 JAV17147.1 KA647450 AFP62079.1 GDHF01009138 JAI43176.1 GBYB01013203 JAG82970.1 GAKP01018685 GAKP01018684 JAC40267.1 GAPW01002460 JAC11138.1 CH916369 EDV92933.1 GEFH01002017 JAP66564.1 CH902623 EDV30193.1 CM000070 EAL29124.1 CH479185 EDW38450.1 CH940650 EDW68423.1 KQ761246 OAD57934.1 CH933806 EDW15318.1 JXJN01013365 OUUW01000008 SPP83849.1 DS231997 EDS30985.1 CP012526 ALC47877.1 CH477489 EAT40089.1
KQ460651 KPJ13039.1 KQ459582 KPI99037.1 NWSH01000176 PCG78732.1 KZ149935 PZC77186.1 AK404038 BAM20112.1 ODYU01001242 SOQ37186.1 NNAY01000986 OXU25605.1 PYGN01000374 PSN47461.1 MH365706 QBB01515.1 GL445685 EFN89233.1 GL765434 EFZ16256.1 GFDG01000570 JAV18229.1 KQ979608 KYN20488.1 GL438820 EFN68392.1 KQ978081 KYM97140.1 LBMM01002952 KMQ94145.1 FX983743 BAX07242.1 NEVH01017450 PNF24298.1 GL888243 EGI64085.1 GDJX01020827 JAT47109.1 DQ062223 AAY63978.1 JRES01001197 KNC24655.1 UFQS01000662 UFQT01000490 UFQT01000662 SSX05893.1 SSX24725.1 GEZM01087903 JAV58405.1 KQ971346 EFA04972.1 KK107139 EZA57723.1 KQ434960 KZC12660.1 KY661311 AVA17398.1 GANO01001560 JAB58311.1 GDAI01001597 JAI16006.1 ADTU01023767 ATLV01015785 KE525036 KFB40725.1 APGK01019088 KB740092 KB632429 ENN81433.1 ERL95462.1 KU932349 APA33985.1 BT128060 AEE63021.1 AXCM01001692 AXCN02000191 GDIQ01191327 GDIP01043456 GDIQ01037520 LRGB01000626 JAK60398.1 JAM60259.1 KZS17437.1 GECU01005761 JAT01946.1 GECZ01025697 JAS44072.1 AAAB01008987 EAA01572.4 APCN01002072 GDIQ01270961 JAJ80763.1 GBXI01004828 JAD09464.1 GEBQ01017166 JAT22811.1 KQ982944 KYQ49026.1 GDIQ01178465 JAK73260.1 GAMD01003320 JAA98270.1 GGFK01001594 MBW34915.1 GGFJ01007143 MBW56284.1 ADMH02002134 GGFL01004373 ETN58378.1 MBW68551.1 GGFL01004365 MBW68543.1 GAMC01003193 JAC03363.1 GGFJ01007144 MBW56285.1 GDIQ01082614 JAN12123.1 KF647639 AHB50501.1 GFDG01001643 JAV17156.1 GGFM01000967 MBW21718.1 CVRI01000051 CRK99511.1 GGFM01000971 MBW21722.1 GL732643 EFX69032.1 JXUM01076841 JXUM01076842 KQ562972 KXJ74736.1 GFDG01001652 JAV17147.1 KA647450 AFP62079.1 GDHF01009138 JAI43176.1 GBYB01013203 JAG82970.1 GAKP01018685 GAKP01018684 JAC40267.1 GAPW01002460 JAC11138.1 CH916369 EDV92933.1 GEFH01002017 JAP66564.1 CH902623 EDV30193.1 CM000070 EAL29124.1 CH479185 EDW38450.1 CH940650 EDW68423.1 KQ761246 OAD57934.1 CH933806 EDW15318.1 JXJN01013365 OUUW01000008 SPP83849.1 DS231997 EDS30985.1 CP012526 ALC47877.1 CH477489 EAT40089.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000053240
UP000053268
UP000218220
+ More
UP000215335 UP000002358 UP000245037 UP000008237 UP000078492 UP000000311 UP000078542 UP000036403 UP000235965 UP000007755 UP000037069 UP000007266 UP000053097 UP000076502 UP000075880 UP000005205 UP000075885 UP000030765 UP000019118 UP000030742 UP000075884 UP000075883 UP000075900 UP000075886 UP000075920 UP000076858 UP000075901 UP000076407 UP000075903 UP000075902 UP000075882 UP000007062 UP000075840 UP000075809 UP000075881 UP000000673 UP000069272 UP000183832 UP000000305 UP000069940 UP000249989 UP000095300 UP000095301 UP000001070 UP000092443 UP000007801 UP000001819 UP000008744 UP000008792 UP000009192 UP000092460 UP000268350 UP000002320 UP000092553 UP000008820
UP000215335 UP000002358 UP000245037 UP000008237 UP000078492 UP000000311 UP000078542 UP000036403 UP000235965 UP000007755 UP000037069 UP000007266 UP000053097 UP000076502 UP000075880 UP000005205 UP000075885 UP000030765 UP000019118 UP000030742 UP000075884 UP000075883 UP000075900 UP000075886 UP000075920 UP000076858 UP000075901 UP000076407 UP000075903 UP000075902 UP000075882 UP000007062 UP000075840 UP000075809 UP000075881 UP000000673 UP000069272 UP000183832 UP000000305 UP000069940 UP000249989 UP000095300 UP000095301 UP000001070 UP000092443 UP000007801 UP000001819 UP000008744 UP000008792 UP000009192 UP000092460 UP000268350 UP000002320 UP000092553 UP000008820
Interpro
Gene 3D
ProteinModelPortal
H9JU51
A0A3S2NCY2
A0A212FMW7
A0A0N1IFM5
A0A194Q0C9
A0A2A4K4Z0
+ More
A0A2W1BWY1 I4DQC2 A0A2H1V8P8 A0A232F4J8 K7J833 A0A2P8YT87 A0A411G6K4 E2B525 E9IT62 A0A1L8EI00 A0A195E5P6 E2ADN0 A0A195C8W5 A0A0J7NNS3 A0A2Z5RE49 A0A2J7Q6U6 F4WPA4 A0A1D1XXI6 Q4PP90 A0A0L0BXC1 A0A336KS61 A0A1Y1KFL2 D2A663 A0A026WP45 A0A154PN63 A0A2P0XJ03 U5EL58 A0A182JME3 A0A0K8TPD6 A0A158NR67 A0A182PC32 A0A084VRY1 N6TTQ3 A0A1I9WLG5 J3JXT9 A0A182NZ02 A0A182MFG3 A0A182R1J5 A0A182QV09 A0A182VQY7 A0A0P6AFG0 A0A1B6JRY2 A0A1B6F1E5 A0A182S6J7 A0A2Y9D3K4 A0A182UXV3 A0A182U2M5 A0A182KM35 Q7PYE7 A0A182IIP6 A0A0P5EE84 A0A0A1XDJ4 A0A1B6LGI7 A0A151WME8 A0A182JQR4 A0A0P5L8R4 T1DMZ1 A0A2M4A2E0 A0A2M4BTF4 W5J4N2 A0A182FQJ7 A0A2M4CT91 W8BVV6 A0A2M4BT81 A0A0P6DCQ6 V5SJ21 A0A1L8EEQ2 A0A2M3YZS5 A0A1J1IGX3 A0A2M3YZM4 E9HGX0 A0A182GNQ1 A0A1L8EEK1 A0A1I8PUI8 T1PH40 A0A0K8VWB8 A0A0C9QFA6 A0A034VDU0 A0A023EQ02 B4JI52 A0A1S4FIY1 A0A1A9YNS9 A0A131XGD2 B3MTR2 Q293U0 B4GLM6 B4M0W4 A0A310SPT5 B4K7H4 A0A1B0BFC4 A0A3B0KCD1 B0WMC8 A0A0M4EPV2 Q16ZI5
A0A2W1BWY1 I4DQC2 A0A2H1V8P8 A0A232F4J8 K7J833 A0A2P8YT87 A0A411G6K4 E2B525 E9IT62 A0A1L8EI00 A0A195E5P6 E2ADN0 A0A195C8W5 A0A0J7NNS3 A0A2Z5RE49 A0A2J7Q6U6 F4WPA4 A0A1D1XXI6 Q4PP90 A0A0L0BXC1 A0A336KS61 A0A1Y1KFL2 D2A663 A0A026WP45 A0A154PN63 A0A2P0XJ03 U5EL58 A0A182JME3 A0A0K8TPD6 A0A158NR67 A0A182PC32 A0A084VRY1 N6TTQ3 A0A1I9WLG5 J3JXT9 A0A182NZ02 A0A182MFG3 A0A182R1J5 A0A182QV09 A0A182VQY7 A0A0P6AFG0 A0A1B6JRY2 A0A1B6F1E5 A0A182S6J7 A0A2Y9D3K4 A0A182UXV3 A0A182U2M5 A0A182KM35 Q7PYE7 A0A182IIP6 A0A0P5EE84 A0A0A1XDJ4 A0A1B6LGI7 A0A151WME8 A0A182JQR4 A0A0P5L8R4 T1DMZ1 A0A2M4A2E0 A0A2M4BTF4 W5J4N2 A0A182FQJ7 A0A2M4CT91 W8BVV6 A0A2M4BT81 A0A0P6DCQ6 V5SJ21 A0A1L8EEQ2 A0A2M3YZS5 A0A1J1IGX3 A0A2M3YZM4 E9HGX0 A0A182GNQ1 A0A1L8EEK1 A0A1I8PUI8 T1PH40 A0A0K8VWB8 A0A0C9QFA6 A0A034VDU0 A0A023EQ02 B4JI52 A0A1S4FIY1 A0A1A9YNS9 A0A131XGD2 B3MTR2 Q293U0 B4GLM6 B4M0W4 A0A310SPT5 B4K7H4 A0A1B0BFC4 A0A3B0KCD1 B0WMC8 A0A0M4EPV2 Q16ZI5
PDB
2DFD
E-value=1.74789e-100,
Score=934
Ontologies
KEGG
PATHWAY
00020
Citrate cycle (TCA cycle) - Bombyx mori (domestic silkworm)
00270 Cysteine and methionine metabolism - Bombyx mori (domestic silkworm)
00620 Pyruvate metabolism - Bombyx mori (domestic silkworm)
00630 Glyoxylate and dicarboxylate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
00270 Cysteine and methionine metabolism - Bombyx mori (domestic silkworm)
00620 Pyruvate metabolism - Bombyx mori (domestic silkworm)
00630 Glyoxylate and dicarboxylate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
GO
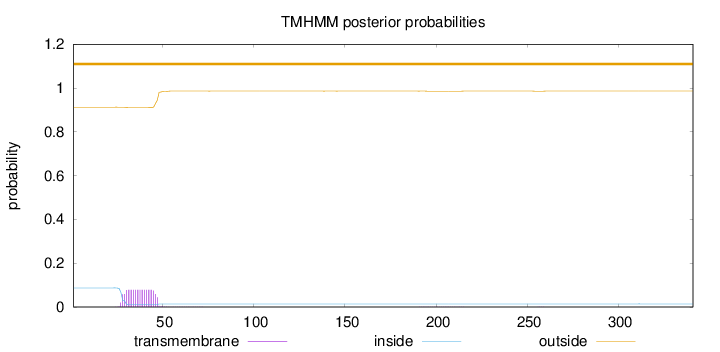
Topology
Length:
341
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.52991
Exp number, first 60 AAs:
1.50838
Total prob of N-in:
0.08734
outside
1 - 341
Population Genetic Test Statistics
Pi
59.606838
Theta
53.850191
Tajima's D
-0.680244
CLR
2273.954092
CSRT
0.199890005499725
Interpretation
Possibly Positive selection
