Gene
KWMTBOMO09470
Pre Gene Modal
BGIBMGA013783
Annotation
AF461149_1_transposase_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.596
Sequence
CDS
ATGGACCTGAGACTGAAGAGATGCCGCGCTTTGTTGAAGCGGTACGCGGGAAAAAAATATCGGGAAATTCTTTTTTCGGATGACAAAATTTTTACCGCAGAAGAGAGCTACAACAAACAAAATGATAAGGTGTACGCACACAGTAGTGAAGAAGCGAGCAACCGTATTCCGCGTGTCCAACGAGGTCATTTTCCATCCTCGCTCATGGTATGGTTGGGAGTTTCTTATTGGGGCTTAACAGAGGTACATTTTTGTGAGAAAGGTGTAAAAACGAATACAGTTGTGTATCAAAACACAGTCCTGACGAACCTTGTGGAACCTGTTTCTCATACCATGTTCAATAACAGGCACTGGGTATTCCAACAAGATTCGGCGCCAGCTCATAGAGCGAAGAGCACACAAGACTGGCTGGCGGCGCGTGAAATCGACCTCATCCGGCACGAAGACTGGCCCTCCTCCAGTTCAGATTTGAATCCGTTAGATTACAAGATATGGCAACATTTGGAGGAAAAGGCGTGCTCAAAGCCTCATCCCAATTTGGAGTCACTCAAGACATCCTTGATTAAGGCAGCCGCCGATATTGACATGGACCTCGTTCGTGCTGCAATAGACGACTGGCCGCGCAGATTGAAAGCCTGTATTCAAAATCATTGA
Protein
MDLRLKRCRALLKRYAGKKYREILFSDDKIFTAEESYNKQNDKVYAHSSEEASNRIPRVQRGHFPSSLMVWLGVSYWGLTEVHFCEKGVKTNTVVYQNTVLTNLVEPVSHTMFNNRHWVFQQDSAPAHRAKSTQDWLAAREIDLIRHEDWPSSSSDLNPLDYKIWQHLEEKACSKPHPNLESLKTSLIKAAADIDMDLVRAAIDDWPRRLKACIQNH
Summary
Similarity
Belongs to the TGF-beta family.
Belongs to the UDP-glycosyltransferase family.
Belongs to the UDP-glycosyltransferase family.
Uniprot
Q8ITJ9
B1Q3L9
B1Q3L5
B9A8E5
B1Q3L8
A0A437BDC1
+ More
A0A3S2LTJ6 A0A437ASV4 A0A3S2M6Z0 A0A437B2G3 A0A3S2LWW4 A0A0N0PF55 A0A3S2NF68 A0A437BVA4 P90693 G3G3K1 A0A3S2M6X4 A0A226D9B6 A0A437B620 A0A437B0M5 A0A1I7SJQ8 A0A3S2LQG1 A0A016TV39 A0A0N4XXA5 A0A368FVD9 A0A1I7SRX5 A0A2G5T9N6 A0A1I7T5D4 A0A2G5SF52 A0A368H7I1 E3NJL6 A0A016S9F8 A0A1I7RU52 A0A1I8CC25 E3N6E8 A0A016TQR4 E3N9D7 B6IKQ6 A0A183F5L1 A0A3P7TF39 W6NUW0 E3MXK7 E3M897 H3EXA9 E3N1H1 E3ND45 H3E4E9 E3NHK7 E3N4H8 A0A016S0M1 A0A016W3L5 E3M1J2 E3NDP6 A0A016VJ32 A0A016WKB6 E3NHD7 E3MXK8 H3EE07 A0A2G5SZY0 E3MQK2 A0A2G5TN79 A0A0N4Y502 A0A016WN34 E3N749 H3E1D2 A0A2G5TSG2 A0A0N4XTP7 A0A2G5VIT1 H3DS69 A0A016WY15 A0A2H2IVX6 A0A2H2I0K4 H3ESY7 A0A2H2J0T6 A0A1I7TL85 A0A2G5TZM3 A0A0K9NHE3 H3FF09 A0A2G5TZ20 H3DZR2 H3F5A8
A0A3S2LTJ6 A0A437ASV4 A0A3S2M6Z0 A0A437B2G3 A0A3S2LWW4 A0A0N0PF55 A0A3S2NF68 A0A437BVA4 P90693 G3G3K1 A0A3S2M6X4 A0A226D9B6 A0A437B620 A0A437B0M5 A0A1I7SJQ8 A0A3S2LQG1 A0A016TV39 A0A0N4XXA5 A0A368FVD9 A0A1I7SRX5 A0A2G5T9N6 A0A1I7T5D4 A0A2G5SF52 A0A368H7I1 E3NJL6 A0A016S9F8 A0A1I7RU52 A0A1I8CC25 E3N6E8 A0A016TQR4 E3N9D7 B6IKQ6 A0A183F5L1 A0A3P7TF39 W6NUW0 E3MXK7 E3M897 H3EXA9 E3N1H1 E3ND45 H3E4E9 E3NHK7 E3N4H8 A0A016S0M1 A0A016W3L5 E3M1J2 E3NDP6 A0A016VJ32 A0A016WKB6 E3NHD7 E3MXK8 H3EE07 A0A2G5SZY0 E3MQK2 A0A2G5TN79 A0A0N4Y502 A0A016WN34 E3N749 H3E1D2 A0A2G5TSG2 A0A0N4XTP7 A0A2G5VIT1 H3DS69 A0A016WY15 A0A2H2IVX6 A0A2H2I0K4 H3ESY7 A0A2H2J0T6 A0A1I7TL85 A0A2G5TZM3 A0A0K9NHE3 H3FF09 A0A2G5TZ20 H3DZR2 H3F5A8
EMBL
AF461149
AAN06610.1
AB363028
BAG15927.1
AB363006
AB363010
+ More
AB363014 BAG15923.1 BAG15924.1 AB473770 BAH20555.1 AB363018 BAG15926.1 RSAL01000084 RVE48403.1 RSAL01000012 RVE53510.1 RSAL01001770 RVE40705.1 RSAL01000021 RVE52408.1 RSAL01000191 RVE44681.1 RSAL01000140 RVE46091.1 KQ459680 KPJ20472.1 RSAL01000063 RVE49524.1 RSAL01000006 RVE54275.1 U47917 AAB47739.1 JF779677 AEO90418.1 RSAL01000022 RVE52367.1 LNIX01000030 OXA41211.1 RSAL01000149 RVE45804.1 RSAL01000236 RVE43715.1 RSAL01000470 RVE41612.1 JARK01001411 EYC06502.1 UYSL01019905 VDL71189.1 JOJR01000583 RCN36166.1 PDUG01000005 PIC23776.1 PDUG01000012 PIC13506.1 JOJR01000015 RCN51257.1 DS268747 EFP00909.1 JARK01001601 EYB87313.1 DS268539 EFO88027.1 JARK01001418 EYC05389.1 DS268565 EFO90304.1 HE601413 CAS00486.1 UZAH01001595 VDO19836.1 CAVP010059404 CDL95787.1 DS268492 EFP11689.1 DS268428 EFO94379.1 DS268508 EFO83264.1 DS268606 EFO93488.1 DS268679 EFO98285.1 DS268525 EFO85497.1 JARK01001658 EYB84158.1 JARK01001337 EYC34439.1 DS268421 EFO88618.1 DS268612 EFO94085.1 JARK01001345 EYC27589.1 JARK01000255 EYC39473.1 DS268673 EFO98001.1 EFP11674.1 PDUG01000006 PIC20538.1 DS268466 EFP06959.1 PIC28734.1 UYSL01020440 VDL74616.1 JARK01000176 EYC41234.1 DS268545 EFO88382.1 PIC29926.1 UYSL01019770 VDL69590.1 PDUG01000001 PIC51673.1 JARK01000059 EYC44525.1 PDUG01000004 PIC32571.1 LFYR01002220 KMZ56176.1 PIC32570.1
AB363014 BAG15923.1 BAG15924.1 AB473770 BAH20555.1 AB363018 BAG15926.1 RSAL01000084 RVE48403.1 RSAL01000012 RVE53510.1 RSAL01001770 RVE40705.1 RSAL01000021 RVE52408.1 RSAL01000191 RVE44681.1 RSAL01000140 RVE46091.1 KQ459680 KPJ20472.1 RSAL01000063 RVE49524.1 RSAL01000006 RVE54275.1 U47917 AAB47739.1 JF779677 AEO90418.1 RSAL01000022 RVE52367.1 LNIX01000030 OXA41211.1 RSAL01000149 RVE45804.1 RSAL01000236 RVE43715.1 RSAL01000470 RVE41612.1 JARK01001411 EYC06502.1 UYSL01019905 VDL71189.1 JOJR01000583 RCN36166.1 PDUG01000005 PIC23776.1 PDUG01000012 PIC13506.1 JOJR01000015 RCN51257.1 DS268747 EFP00909.1 JARK01001601 EYB87313.1 DS268539 EFO88027.1 JARK01001418 EYC05389.1 DS268565 EFO90304.1 HE601413 CAS00486.1 UZAH01001595 VDO19836.1 CAVP010059404 CDL95787.1 DS268492 EFP11689.1 DS268428 EFO94379.1 DS268508 EFO83264.1 DS268606 EFO93488.1 DS268679 EFO98285.1 DS268525 EFO85497.1 JARK01001658 EYB84158.1 JARK01001337 EYC34439.1 DS268421 EFO88618.1 DS268612 EFO94085.1 JARK01001345 EYC27589.1 JARK01000255 EYC39473.1 DS268673 EFO98001.1 EFP11674.1 PDUG01000006 PIC20538.1 DS268466 EFP06959.1 PIC28734.1 UYSL01020440 VDL74616.1 JARK01000176 EYC41234.1 DS268545 EFO88382.1 PIC29926.1 UYSL01019770 VDL69590.1 PDUG01000001 PIC51673.1 JARK01000059 EYC44525.1 PDUG01000004 PIC32571.1 LFYR01002220 KMZ56176.1 PIC32570.1
Proteomes
Pfam
Interpro
IPR009057
Homeobox-like_sf
+ More
IPR001839 TGF-b_C
IPR029034 Cystine-knot_cytokine
IPR017948 TGFb_CS
IPR015615 TGF-beta-rel
IPR036259 MFS_trans_sf
IPR032171 COR
IPR020859 ROC_dom
IPR000719 Prot_kinase_dom
IPR011047 Quinoprotein_ADH-like_supfam
IPR011009 Kinase-like_dom_sf
IPR008271 Ser/Thr_kinase_AS
IPR027417 P-loop_NTPase
IPR038717 Tc1-like_DDE_dom
IPR002213 UDP_glucos_trans
IPR035595 UDP_glycos_trans_CS
IPR036388 WH-like_DNA-bd_sf
IPR001523 Paired_dom
IPR036906 ATPase_V1_fsu_sf
IPR036390 WH_DNA-bd_sf
IPR001766 Fork_head_dom
IPR030456 TF_fork_head_CS_2
IPR001839 TGF-b_C
IPR029034 Cystine-knot_cytokine
IPR017948 TGFb_CS
IPR015615 TGF-beta-rel
IPR036259 MFS_trans_sf
IPR032171 COR
IPR020859 ROC_dom
IPR000719 Prot_kinase_dom
IPR011047 Quinoprotein_ADH-like_supfam
IPR011009 Kinase-like_dom_sf
IPR008271 Ser/Thr_kinase_AS
IPR027417 P-loop_NTPase
IPR038717 Tc1-like_DDE_dom
IPR002213 UDP_glucos_trans
IPR035595 UDP_glycos_trans_CS
IPR036388 WH-like_DNA-bd_sf
IPR001523 Paired_dom
IPR036906 ATPase_V1_fsu_sf
IPR036390 WH_DNA-bd_sf
IPR001766 Fork_head_dom
IPR030456 TF_fork_head_CS_2
SUPFAM
Gene 3D
CDD
ProteinModelPortal
Q8ITJ9
B1Q3L9
B1Q3L5
B9A8E5
B1Q3L8
A0A437BDC1
+ More
A0A3S2LTJ6 A0A437ASV4 A0A3S2M6Z0 A0A437B2G3 A0A3S2LWW4 A0A0N0PF55 A0A3S2NF68 A0A437BVA4 P90693 G3G3K1 A0A3S2M6X4 A0A226D9B6 A0A437B620 A0A437B0M5 A0A1I7SJQ8 A0A3S2LQG1 A0A016TV39 A0A0N4XXA5 A0A368FVD9 A0A1I7SRX5 A0A2G5T9N6 A0A1I7T5D4 A0A2G5SF52 A0A368H7I1 E3NJL6 A0A016S9F8 A0A1I7RU52 A0A1I8CC25 E3N6E8 A0A016TQR4 E3N9D7 B6IKQ6 A0A183F5L1 A0A3P7TF39 W6NUW0 E3MXK7 E3M897 H3EXA9 E3N1H1 E3ND45 H3E4E9 E3NHK7 E3N4H8 A0A016S0M1 A0A016W3L5 E3M1J2 E3NDP6 A0A016VJ32 A0A016WKB6 E3NHD7 E3MXK8 H3EE07 A0A2G5SZY0 E3MQK2 A0A2G5TN79 A0A0N4Y502 A0A016WN34 E3N749 H3E1D2 A0A2G5TSG2 A0A0N4XTP7 A0A2G5VIT1 H3DS69 A0A016WY15 A0A2H2IVX6 A0A2H2I0K4 H3ESY7 A0A2H2J0T6 A0A1I7TL85 A0A2G5TZM3 A0A0K9NHE3 H3FF09 A0A2G5TZ20 H3DZR2 H3F5A8
A0A3S2LTJ6 A0A437ASV4 A0A3S2M6Z0 A0A437B2G3 A0A3S2LWW4 A0A0N0PF55 A0A3S2NF68 A0A437BVA4 P90693 G3G3K1 A0A3S2M6X4 A0A226D9B6 A0A437B620 A0A437B0M5 A0A1I7SJQ8 A0A3S2LQG1 A0A016TV39 A0A0N4XXA5 A0A368FVD9 A0A1I7SRX5 A0A2G5T9N6 A0A1I7T5D4 A0A2G5SF52 A0A368H7I1 E3NJL6 A0A016S9F8 A0A1I7RU52 A0A1I8CC25 E3N6E8 A0A016TQR4 E3N9D7 B6IKQ6 A0A183F5L1 A0A3P7TF39 W6NUW0 E3MXK7 E3M897 H3EXA9 E3N1H1 E3ND45 H3E4E9 E3NHK7 E3N4H8 A0A016S0M1 A0A016W3L5 E3M1J2 E3NDP6 A0A016VJ32 A0A016WKB6 E3NHD7 E3MXK8 H3EE07 A0A2G5SZY0 E3MQK2 A0A2G5TN79 A0A0N4Y502 A0A016WN34 E3N749 H3E1D2 A0A2G5TSG2 A0A0N4XTP7 A0A2G5VIT1 H3DS69 A0A016WY15 A0A2H2IVX6 A0A2H2I0K4 H3ESY7 A0A2H2J0T6 A0A1I7TL85 A0A2G5TZM3 A0A0K9NHE3 H3FF09 A0A2G5TZ20 H3DZR2 H3F5A8
PDB
5CR4
E-value=3.61488e-09,
Score=144
Ontologies
GO
PANTHER
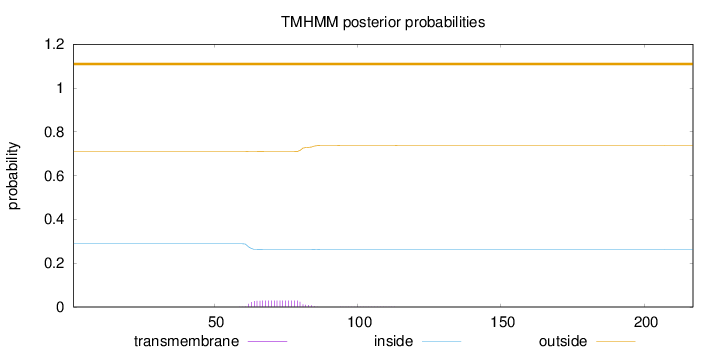
Topology
Subcellular location
Secreted
Nucleus
Nucleus
Length:
217
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.578099999999999
Exp number, first 60 AAs:
0.00082
Total prob of N-in:
0.28903
outside
1 - 217
Population Genetic Test Statistics
Pi
59.316139
Theta
38.54625
Tajima's D
1.479317
CLR
0.320454
CSRT
0.781160941952902
Interpretation
Uncertain
