Gene
KWMTBOMO09467 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013070
Annotation
PREDICTED:_hsp70-binding_protein_1_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.674 Extracellular Reliability : 1.475 Nuclear Reliability : 1.152
Sequence
CDS
ATGAGTTCGAATAATACGCAGAATAACACGATTGCCGGAGCTCTGACTTATCCGACAAGAAGTCAAGATGAAGTCCTACCAAATCAGCCGAGGCAGCCAAGAAACCTGCAAGGACTTTTACGGTTTGCAATGGAAGCCACAAAAGCTGAAGATGCACCAGGGAACTCGGAATTAGGACCAATTGATGATGAAAGACGCAAATTCCTTGAAGACGCCCTAAAGAGTCTGACAGTGAACATTGCTGAAGTGTTACTGAACGCCATCAGAATTCTCACCAATACTGAAAGGATTCGTAGCATACAGTTTGGGGAACCACTACCAGAAGACGTACAGGCGGCTTTTGACAATGTCCTTGAATATATTGACGATATTGATACAGCAAATGACTTCTATAAGATGGGAGGGTTTGCTATGTTTCCAATATGCTATGGAAGCGAAAATGAGAAGGTCCGTGCCCTCGCCAGTACGGTCCTGGCTGAGCTCTGTCAGAACAATCCGTTCTGCCAGGAGAAGTTCTTAGAGTACGGTCTGCTGAACGTTCTACTCACTCTAGTGCAATCAGAGCAAGGCGAAGCCCTCGTCAAGTGCCTATATGCTTTATCATGCGGTTGCCGCGAGTACTCGCCCGCGTGTCGCTCGCTGGTGTCGCGCGGGGGGTGCGGGGCGTTGGGGGCGGCGCTGGCGCAGGACGAGCCCTCCGCCAGGACCAAGGCCGCCTTCCTCATCAGGTACCTGGCCAGCCAATACAGCGACGCTAAAGAGCAGTTCATCCAGCACAATGTAGTCAGGACCATAGCTGAACAACTGAGAGGGGGTCGTGACGCGACCAGTGAGCACTTCCTGAACTTACTCTGCACTCTGCTCGAGGACCTGGACCCCAGGGTCCTGGACCAATGCCGGGACCCCACCCTACACTTGAAGGAGACTTTAGAACAACACTTAGGACACCCTGAGCTGGAAAGTTTTACGGAAGAAAAGGAGTATTGTGAAGAGATCCTCAGGAAGGTGTTTGTCGGGCATAAGCCAGTGGAGATCGTGGTTGAAGAAGCCGATAGATAG
Protein
MSSNNTQNNTIAGALTYPTRSQDEVLPNQPRQPRNLQGLLRFAMEATKAEDAPGNSELGPIDDERRKFLEDALKSLTVNIAEVLLNAIRILTNTERIRSIQFGEPLPEDVQAAFDNVLEYIDDIDTANDFYKMGGFAMFPICYGSENEKVRALASTVLAELCQNNPFCQEKFLEYGLLNVLLTLVQSEQGEALVKCLYALSCGCREYSPACRSLVSRGGCGALGAALAQDEPSARTKAAFLIRYLASQYSDAKEQFIQHNVVRTIAEQLRGGRDATSEHFLNLLCTLLEDLDPRVLDQCRDPTLHLKETLEQHLGHPELESFTEEKEYCEEILRKVFVGHKPVEIVVEEADR
Summary
Similarity
Belongs to the archaeal rpoM/eukaryotic RPA12/RPB9/RPC11 RNA polymerase family.
Uniprot
H9JU58
S4PMK3
A0A437BC08
A0A212FMW3
A0A2A4K4A3
A0A2W1BWZ2
+ More
A0A0N0PC63 I4DKG5 A0A1Y1N9D1 K7IM62 D6X1Z2 A0A0T6B7Z3 A0A1W4X300 A0A411G6F2 A0A154PA11 V5G7K4 A0A195DDV7 A0A2J7QZD9 A0A0L7RDC2 A0A232EZB7 E1ZYB3 A0A195FUD3 A0A0N0U2Z3 U5EHS2 A0A158P457 A0A195C632 A0A087ZNK4 A0A195BD66 Q17K40 A0A182H8P6 A0A1S4F003 A0A023ENC3 A0A023ENB3 A0A182QC69 Q17K39 A0A182XFX1 A0A182TFV7 A0A182L0I5 Q7PN96 A0A182IC56 A0A1S4EZU1 A0A2A3E3S7 A0A067QRK3 A0A182VJL0 A0A151X9Z8 A0A084WH63 J3JWM2 A0A182NU06 A0A182KG39 A0A182S8X8 A0A182JEQ4 A0A182PLS3 A0A026WYI9 A0A182YDT5 R4WDY6 A0A1Q3FED5 E2B4J1 A0A2U9IAV0 A0A1S4J085 A0A1J1I678 A0A0P5G7V8 E0VPZ3 A0A336MNE5 A0A0P6I5D6 A0A0N8AXI8 A0A1E1XA21 A0A0P4X0P1 A0A023GIJ3 A0A433U8V5 A0A0A9XFZ8 A0A0K8TDS5 A0A0B8RPC7 A0A0N8AIG0 A0A0P4XIJ2 A0A2R7WD43 B0VZZ3 A0A0P4X077 E9J2L1 A0A1E1XR90 A0A0J7K3L2 A0A3Q0JIB5 A0A131YA91 V5HP71 A0A131XHA3 A0A224XNC0
A0A0N0PC63 I4DKG5 A0A1Y1N9D1 K7IM62 D6X1Z2 A0A0T6B7Z3 A0A1W4X300 A0A411G6F2 A0A154PA11 V5G7K4 A0A195DDV7 A0A2J7QZD9 A0A0L7RDC2 A0A232EZB7 E1ZYB3 A0A195FUD3 A0A0N0U2Z3 U5EHS2 A0A158P457 A0A195C632 A0A087ZNK4 A0A195BD66 Q17K40 A0A182H8P6 A0A1S4F003 A0A023ENC3 A0A023ENB3 A0A182QC69 Q17K39 A0A182XFX1 A0A182TFV7 A0A182L0I5 Q7PN96 A0A182IC56 A0A1S4EZU1 A0A2A3E3S7 A0A067QRK3 A0A182VJL0 A0A151X9Z8 A0A084WH63 J3JWM2 A0A182NU06 A0A182KG39 A0A182S8X8 A0A182JEQ4 A0A182PLS3 A0A026WYI9 A0A182YDT5 R4WDY6 A0A1Q3FED5 E2B4J1 A0A2U9IAV0 A0A1S4J085 A0A1J1I678 A0A0P5G7V8 E0VPZ3 A0A336MNE5 A0A0P6I5D6 A0A0N8AXI8 A0A1E1XA21 A0A0P4X0P1 A0A023GIJ3 A0A433U8V5 A0A0A9XFZ8 A0A0K8TDS5 A0A0B8RPC7 A0A0N8AIG0 A0A0P4XIJ2 A0A2R7WD43 B0VZZ3 A0A0P4X077 E9J2L1 A0A1E1XR90 A0A0J7K3L2 A0A3Q0JIB5 A0A131YA91 V5HP71 A0A131XHA3 A0A224XNC0
Pubmed
19121390
23622113
22118469
28756777
26354079
22651552
+ More
28004739 20075255 18362917 19820115 28648823 20798317 21347285 17510324 26483478 24945155 20966253 12364791 14747013 17210077 24845553 24438588 22516182 23537049 24508170 30249741 25244985 23691247 20566863 28503490 25401762 26823975 21282665 29209593 25765539 28049606
28004739 20075255 18362917 19820115 28648823 20798317 21347285 17510324 26483478 24945155 20966253 12364791 14747013 17210077 24845553 24438588 22516182 23537049 24508170 30249741 25244985 23691247 20566863 28503490 25401762 26823975 21282665 29209593 25765539 28049606
EMBL
BABH01004119
GAIX01003510
JAA89050.1
RSAL01000093
RVE47903.1
AGBW02007651
+ More
OWR55092.1 NWSH01000176 PCG78738.1 KZ149935 PZC77196.1 KQ460651 KPJ13047.1 AK401783 KQ459582 BAM18405.1 KPI99029.1 GEZM01014118 JAV92197.1 KQ971371 EFA09946.1 LJIG01009263 KRT83430.1 MH365661 QBB01470.1 KQ434856 KZC08673.1 GALX01002351 JAB66115.1 KQ980989 KYN10604.1 NEVH01009070 PNF33925.1 KQ414614 KOC68800.1 NNAY01001594 OXU23488.1 GL435204 EFN73812.1 KQ981264 KYN44031.1 KQ438551 KOX67201.1 GANO01002904 JAB56967.1 ADTU01001346 KQ978220 KYM96307.1 KQ976519 KYM82152.1 CH477228 EAT47053.1 JXUM01118898 KQ566235 KXJ70295.1 GAPW01002696 JAC10902.1 GAPW01002695 JAC10903.1 AXCN02000808 EAT47052.1 AAAB01008964 EAA12545.5 APCN01000775 KZ288427 PBC25906.1 KK853042 KDR12144.1 KQ982353 KYQ57202.1 ATLV01023791 KE525346 KFB49557.1 APGK01019170 BT127640 KB740092 KB632310 AEE62602.1 ENN81479.1 ERL92102.1 KK107064 QOIP01000008 EZA60831.1 RLU19169.1 AK417974 BAN21189.1 GFDL01009136 JAV25909.1 GL445565 EFN89391.1 MF797800 AWR93091.1 CVRI01000038 CRK94438.1 GDIQ01246006 LRGB01000944 JAK05719.1 KZS14854.1 DS235379 EEB15449.1 UFQS01001121 UFQT01001121 UFQT01001734 SSX09102.1 SSX31570.1 GDIQ01016625 JAN78112.1 GDIQ01233155 JAK18570.1 GFAC01003084 JAT96104.1 GDIP01249396 JAI74005.1 GBBM01001702 JAC33716.1 RQTK01000037 RUS90247.1 GBHO01023983 GBHO01023982 GBHO01023981 GDHC01006148 GDHC01001932 JAG19621.1 JAG19622.1 JAG19623.1 JAQ12481.1 JAQ16697.1 GBRD01002506 JAG63315.1 GBRD01002507 JAG63314.1 GDIP01144257 JAJ79145.1 GDIP01242563 JAI80838.1 KK854643 PTY17572.1 DS231816 EDS37565.1 GDRN01039609 JAI67186.1 GL767895 EFZ12946.1 GFAA01001602 JAU01833.1 LBMM01015172 KMQ84904.1 GEFM01000371 JAP75425.1 GANP01004704 JAB79764.1 GEFH01003026 JAP65555.1 GFTR01005108 JAW11318.1
OWR55092.1 NWSH01000176 PCG78738.1 KZ149935 PZC77196.1 KQ460651 KPJ13047.1 AK401783 KQ459582 BAM18405.1 KPI99029.1 GEZM01014118 JAV92197.1 KQ971371 EFA09946.1 LJIG01009263 KRT83430.1 MH365661 QBB01470.1 KQ434856 KZC08673.1 GALX01002351 JAB66115.1 KQ980989 KYN10604.1 NEVH01009070 PNF33925.1 KQ414614 KOC68800.1 NNAY01001594 OXU23488.1 GL435204 EFN73812.1 KQ981264 KYN44031.1 KQ438551 KOX67201.1 GANO01002904 JAB56967.1 ADTU01001346 KQ978220 KYM96307.1 KQ976519 KYM82152.1 CH477228 EAT47053.1 JXUM01118898 KQ566235 KXJ70295.1 GAPW01002696 JAC10902.1 GAPW01002695 JAC10903.1 AXCN02000808 EAT47052.1 AAAB01008964 EAA12545.5 APCN01000775 KZ288427 PBC25906.1 KK853042 KDR12144.1 KQ982353 KYQ57202.1 ATLV01023791 KE525346 KFB49557.1 APGK01019170 BT127640 KB740092 KB632310 AEE62602.1 ENN81479.1 ERL92102.1 KK107064 QOIP01000008 EZA60831.1 RLU19169.1 AK417974 BAN21189.1 GFDL01009136 JAV25909.1 GL445565 EFN89391.1 MF797800 AWR93091.1 CVRI01000038 CRK94438.1 GDIQ01246006 LRGB01000944 JAK05719.1 KZS14854.1 DS235379 EEB15449.1 UFQS01001121 UFQT01001121 UFQT01001734 SSX09102.1 SSX31570.1 GDIQ01016625 JAN78112.1 GDIQ01233155 JAK18570.1 GFAC01003084 JAT96104.1 GDIP01249396 JAI74005.1 GBBM01001702 JAC33716.1 RQTK01000037 RUS90247.1 GBHO01023983 GBHO01023982 GBHO01023981 GDHC01006148 GDHC01001932 JAG19621.1 JAG19622.1 JAG19623.1 JAQ12481.1 JAQ16697.1 GBRD01002506 JAG63315.1 GBRD01002507 JAG63314.1 GDIP01144257 JAJ79145.1 GDIP01242563 JAI80838.1 KK854643 PTY17572.1 DS231816 EDS37565.1 GDRN01039609 JAI67186.1 GL767895 EFZ12946.1 GFAA01001602 JAU01833.1 LBMM01015172 KMQ84904.1 GEFM01000371 JAP75425.1 GANP01004704 JAB79764.1 GEFH01003026 JAP65555.1 GFTR01005108 JAW11318.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000218220
UP000053240
UP000053268
+ More
UP000002358 UP000007266 UP000192223 UP000076502 UP000078492 UP000235965 UP000053825 UP000215335 UP000000311 UP000078541 UP000053105 UP000005205 UP000078542 UP000005203 UP000078540 UP000008820 UP000069940 UP000249989 UP000075886 UP000076407 UP000075902 UP000075882 UP000007062 UP000075840 UP000242457 UP000027135 UP000075903 UP000075809 UP000030765 UP000019118 UP000030742 UP000075884 UP000075881 UP000075901 UP000075880 UP000075885 UP000053097 UP000279307 UP000076408 UP000008237 UP000183832 UP000076858 UP000009046 UP000271974 UP000002320 UP000036403 UP000079169
UP000002358 UP000007266 UP000192223 UP000076502 UP000078492 UP000235965 UP000053825 UP000215335 UP000000311 UP000078541 UP000053105 UP000005205 UP000078542 UP000005203 UP000078540 UP000008820 UP000069940 UP000249989 UP000075886 UP000076407 UP000075902 UP000075882 UP000007062 UP000075840 UP000242457 UP000027135 UP000075903 UP000075809 UP000030765 UP000019118 UP000030742 UP000075884 UP000075881 UP000075901 UP000075880 UP000075885 UP000053097 UP000279307 UP000076408 UP000008237 UP000183832 UP000076858 UP000009046 UP000271974 UP000002320 UP000036403 UP000079169
PRIDE
Interpro
SUPFAM
SSF48371
SSF48371
Gene 3D
ProteinModelPortal
H9JU58
S4PMK3
A0A437BC08
A0A212FMW3
A0A2A4K4A3
A0A2W1BWZ2
+ More
A0A0N0PC63 I4DKG5 A0A1Y1N9D1 K7IM62 D6X1Z2 A0A0T6B7Z3 A0A1W4X300 A0A411G6F2 A0A154PA11 V5G7K4 A0A195DDV7 A0A2J7QZD9 A0A0L7RDC2 A0A232EZB7 E1ZYB3 A0A195FUD3 A0A0N0U2Z3 U5EHS2 A0A158P457 A0A195C632 A0A087ZNK4 A0A195BD66 Q17K40 A0A182H8P6 A0A1S4F003 A0A023ENC3 A0A023ENB3 A0A182QC69 Q17K39 A0A182XFX1 A0A182TFV7 A0A182L0I5 Q7PN96 A0A182IC56 A0A1S4EZU1 A0A2A3E3S7 A0A067QRK3 A0A182VJL0 A0A151X9Z8 A0A084WH63 J3JWM2 A0A182NU06 A0A182KG39 A0A182S8X8 A0A182JEQ4 A0A182PLS3 A0A026WYI9 A0A182YDT5 R4WDY6 A0A1Q3FED5 E2B4J1 A0A2U9IAV0 A0A1S4J085 A0A1J1I678 A0A0P5G7V8 E0VPZ3 A0A336MNE5 A0A0P6I5D6 A0A0N8AXI8 A0A1E1XA21 A0A0P4X0P1 A0A023GIJ3 A0A433U8V5 A0A0A9XFZ8 A0A0K8TDS5 A0A0B8RPC7 A0A0N8AIG0 A0A0P4XIJ2 A0A2R7WD43 B0VZZ3 A0A0P4X077 E9J2L1 A0A1E1XR90 A0A0J7K3L2 A0A3Q0JIB5 A0A131YA91 V5HP71 A0A131XHA3 A0A224XNC0
A0A0N0PC63 I4DKG5 A0A1Y1N9D1 K7IM62 D6X1Z2 A0A0T6B7Z3 A0A1W4X300 A0A411G6F2 A0A154PA11 V5G7K4 A0A195DDV7 A0A2J7QZD9 A0A0L7RDC2 A0A232EZB7 E1ZYB3 A0A195FUD3 A0A0N0U2Z3 U5EHS2 A0A158P457 A0A195C632 A0A087ZNK4 A0A195BD66 Q17K40 A0A182H8P6 A0A1S4F003 A0A023ENC3 A0A023ENB3 A0A182QC69 Q17K39 A0A182XFX1 A0A182TFV7 A0A182L0I5 Q7PN96 A0A182IC56 A0A1S4EZU1 A0A2A3E3S7 A0A067QRK3 A0A182VJL0 A0A151X9Z8 A0A084WH63 J3JWM2 A0A182NU06 A0A182KG39 A0A182S8X8 A0A182JEQ4 A0A182PLS3 A0A026WYI9 A0A182YDT5 R4WDY6 A0A1Q3FED5 E2B4J1 A0A2U9IAV0 A0A1S4J085 A0A1J1I678 A0A0P5G7V8 E0VPZ3 A0A336MNE5 A0A0P6I5D6 A0A0N8AXI8 A0A1E1XA21 A0A0P4X0P1 A0A023GIJ3 A0A433U8V5 A0A0A9XFZ8 A0A0K8TDS5 A0A0B8RPC7 A0A0N8AIG0 A0A0P4XIJ2 A0A2R7WD43 B0VZZ3 A0A0P4X077 E9J2L1 A0A1E1XR90 A0A0J7K3L2 A0A3Q0JIB5 A0A131YA91 V5HP71 A0A131XHA3 A0A224XNC0
Ontologies
GO
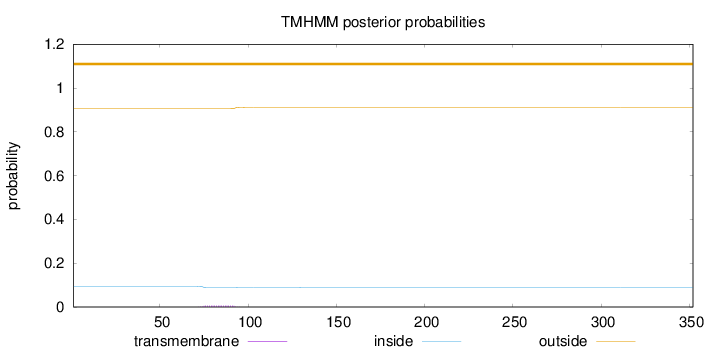
Topology
Length:
352
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.12164
Exp number, first 60 AAs:
0
Total prob of N-in:
0.09429
outside
1 - 352
Population Genetic Test Statistics
Pi
195.90157
Theta
166.432921
Tajima's D
0.688479
CLR
0.875037
CSRT
0.564721763911804
Interpretation
Uncertain
