Gene
KWMTBOMO09465
Pre Gene Modal
BGIBMGA012791
Annotation
Transmembrane_protease_serine_9_[Papilio_xuthus]
Location in the cell
Extracellular Reliability : 2.7
Sequence
CDS
ATGTGGTGGATTTTGATTTGTTCTGTGCTTTTTGGAGGTTCGCAGGGCGTTCCTGTGGAAACTGTAGATGTAATTATACCTTCTGTCCCAGTCCACTTTCAAGATATCCCTCGCAATGACCAAAATCGTATCATAGGCGGATGGCTGGCCACAGAAGGACAGTTCCCTTACAAGGTGAGCGTGCGAAACGTCAGCCCTGTGGGCGGAGTCAGCGCGTGTGGGGGCTCCATAATTCACAACGAATGGATTCTAACAGCAGCTCATTGCGTGGCCAGACGCGAATCATTCGTGATCCGTTTGGGATTGACCAACCTGACCCGTCCTGAGTTAATAATGGAGACAGTGGAGAAGTACGTGCACCCTGACTACAATGTCGGCTCCACCGCCGTGCAGGTTGATGAGATCGCTCTCCTGAAGCTGCCCCGTCGTGTGGAGTATGGACGCGGCTATGTCCCCGAGGTCTTGAGTTGGGTCTATCAATTGGGTATCACGAACGAGGAGTGCCTTAACTGGTACCCTAACAGTCAGACCATCAAGGACCAGACATTGTGCGCGAAATATTATAATCATACTTCACAAAACATTTGTACAGGCGACAGTGGCGGACCCCTGACTATGGATGATCTCGATGGCTCACCCACTATCGTAGGCATCACTAACTTTGGTTACGCGCGGGGATGCAACACACCCAATCCGTCCGGTTTCGTCCGACCAGAGTTCTACCACGACTGGCTGAAACAGGTCACCGGCATCAACTTCGACTGGAGCGTCTTTGACATTGTTATCAGCTAA
Protein
MWWILICSVLFGGSQGVPVETVDVIIPSVPVHFQDIPRNDQNRIIGGWLATEGQFPYKVSVRNVSPVGGVSACGGSIIHNEWILTAAHCVARRESFVIRLGLTNLTRPELIMETVEKYVHPDYNVGSTAVQVDEIALLKLPRRVEYGRGYVPEVLSWVYQLGITNEECLNWYPNSQTIKDQTLCAKYYNHTSQNICTGDSGGPLTMDDLDGSPTIVGITNFGYARGCNTPNPSGFVRPEFYHDWLKQVTGINFDWSVFDIVIS
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
H9JTC9
A0A194Q127
A0A2W1BS57
A0A2A4K4P0
Q9BMQ7
J7FDE1
+ More
A0A0N1IP15 A0A0K8S2W5 A0A2W1BWI0 B4X992 H9JTD0 C9W8H1 A0A0X9I156 A0A2H1V7Z9 A0A2A4K3Y4 A0A437BBW9 C9W8G6 T1QDG4 A0A109P6B5 A0A2H1WBT2 A0A2H1VV25 A0A2A4K3B0 A0PGD9 B6D1R0 E7D010 B6D1Q9 O18437 A0A1E1WQI5 A0A1B0RHN9 A0A1E1WG34 A0A3S2LZV4 A0A2A4K409 A0A2A4K3D4 J7FDV5 A0A2A4K426 Q9XY10 A0A2A4K2C1 A0A2W1BQ29 B1NLE3 A0A2A4K3S1 A0A410KK64 H9JTC5 H9JTC6 A0A2A4K2C9 A0A194Q123 A0A2W1BZC6 A0A2A4K3B1 C9W8I8 A0A212FMW9 A0A2H1VZ50 A0A212F859 C9W8F4 A0A212FMY7 A0A1E1W214 A0A194Q024 A0A194RS93 A0A2H1VQY4 H9JTB5 A0A3S2NJF1 A0A2A4K2D7 Q5MGG6 A0A2H1VX31 A0A2A4K0W2 A0A2H1VXD9 A0A182IVJ9 A0A084VZT5 A0A2Z2GQC5 J7FCI3 A0A1J1HVG8 Q8I8E4 U5EJM0 A0A182G5U5 A0A1Y1LVG1 A0A182G5U6 B0WD46 Q8I8E2 A0A182K8Q1 A0A1Q3FQQ0 A0A1Q3FS52 A0A1Q3FS75 A0A1S4FZ18 Q8I8E3 Q16JM8 Q9GTK7 Q66UD0 B0WD43 A0A1I8NFS9 Q86PL8 A0A182SGP5 Q52NW5 A0A1V1FIT7 B0WD41 D2A3W9 A0A194Q1Y2 Q27440 A0A182MAA4 Q16JN3 Q9NB49
A0A0N1IP15 A0A0K8S2W5 A0A2W1BWI0 B4X992 H9JTD0 C9W8H1 A0A0X9I156 A0A2H1V7Z9 A0A2A4K3Y4 A0A437BBW9 C9W8G6 T1QDG4 A0A109P6B5 A0A2H1WBT2 A0A2H1VV25 A0A2A4K3B0 A0PGD9 B6D1R0 E7D010 B6D1Q9 O18437 A0A1E1WQI5 A0A1B0RHN9 A0A1E1WG34 A0A3S2LZV4 A0A2A4K409 A0A2A4K3D4 J7FDV5 A0A2A4K426 Q9XY10 A0A2A4K2C1 A0A2W1BQ29 B1NLE3 A0A2A4K3S1 A0A410KK64 H9JTC5 H9JTC6 A0A2A4K2C9 A0A194Q123 A0A2W1BZC6 A0A2A4K3B1 C9W8I8 A0A212FMW9 A0A2H1VZ50 A0A212F859 C9W8F4 A0A212FMY7 A0A1E1W214 A0A194Q024 A0A194RS93 A0A2H1VQY4 H9JTB5 A0A3S2NJF1 A0A2A4K2D7 Q5MGG6 A0A2H1VX31 A0A2A4K0W2 A0A2H1VXD9 A0A182IVJ9 A0A084VZT5 A0A2Z2GQC5 J7FCI3 A0A1J1HVG8 Q8I8E4 U5EJM0 A0A182G5U5 A0A1Y1LVG1 A0A182G5U6 B0WD46 Q8I8E2 A0A182K8Q1 A0A1Q3FQQ0 A0A1Q3FS52 A0A1Q3FS75 A0A1S4FZ18 Q8I8E3 Q16JM8 Q9GTK7 Q66UD0 B0WD43 A0A1I8NFS9 Q86PL8 A0A182SGP5 Q52NW5 A0A1V1FIT7 B0WD41 D2A3W9 A0A194Q1Y2 Q27440 A0A182MAA4 Q16JN3 Q9NB49
Pubmed
EMBL
BABH01004122
BABH01004123
BABH01004124
KQ459582
KPI99028.1
KZ149935
+ More
PZC77161.1 NWSH01000176 PCG78733.1 AF309500 AAK06410.1 JN252043 AFM28256.1 KQ460651 KPJ13048.1 GBRD01018206 JAG47621.1 PZC77160.1 EF531628 ABR88239.1 BABH01004119 BABH01004120 BABH01004121 FJ205416 ACR15984.1 KP690084 ALO61086.1 ODYU01001143 SOQ36937.1 PCG78736.1 RSAL01000093 RVE47909.1 FJ205411 ACR15979.2 JQ429439 AFW03964.1 KP690083 ALO61085.1 ODYU01007484 SOQ50292.1 ODYU01004626 SOQ44697.1 PCG78737.1 AY618894 AAV33657.1 EU673453 ACI45416.1 HM990180 ADT80828.1 EU673452 ACI45415.1 Y12272 CAA72951.1 GDQN01001948 JAT89106.1 KM360194 AKH49610.1 GDQN01005223 JAT85831.1 RVE47905.1 NWSH01000207 PCG78410.1 PCG78414.1 JN252040 AFM28253.1 PCG78412.1 AB026735 BAA77401.1 PCG78411.1 PZC77162.1 EF600058 ABU98623.1 PCG78413.1 MH201547 QAR48640.1 BABH01004141 BABH01004139 BABH01004140 PCG78415.1 KPI99023.1 PZC77163.1 PCG78408.1 FJ205435 ACR16001.2 AGBW02007651 OWR55096.1 ODYU01005353 SOQ46109.1 AGBW02009787 OWR49920.1 FJ205398 ACR15967.2 OWR55112.1 GDQN01010016 JAT81038.1 KPI98932.1 KQ459700 KPJ20347.1 ODYU01003737 SOQ42892.1 BABH01004171 BABH01004172 RSAL01000071 RVE49104.1 PCG78407.1 AY829820 AAV91434.2 ODYU01004804 SOQ45062.1 NWSH01000261 PCG77915.1 ODYU01005031 SOQ45511.1 ATLV01018958 KE525256 KFB43479.1 KX951431 ARR75599.1 JN252039 AFM28252.1 CVRI01000022 CRK92071.1 AY155562 AAN75000.1 GANO01002186 JAB57685.1 JXUM01007351 KQ560238 KXJ83665.1 GEZM01046011 JAV77579.1 JXUM01007352 KXJ83666.1 DS231893 EDS44181.1 AY155564 AAN75002.1 GFDL01005094 JAV29951.1 GFDL01004648 JAV30397.1 GFDL01004647 JAV30398.1 AY155563 AAN75001.1 CH478002 EAT34483.1 AF268665 AAG10005.1 AY603563 AAU06478.1 EDS44178.1 AY198134 AAO43403.1 AY995188 AAX94742.1 FX985331 BAX07344.1 EDS44176.1 KQ971348 EFA05747.1 KPI99004.1 M77814 AAA29356.1 AXCM01001005 EAT34478.1 AF266757 AF187078 AAF82286.1 AAG17021.1
PZC77161.1 NWSH01000176 PCG78733.1 AF309500 AAK06410.1 JN252043 AFM28256.1 KQ460651 KPJ13048.1 GBRD01018206 JAG47621.1 PZC77160.1 EF531628 ABR88239.1 BABH01004119 BABH01004120 BABH01004121 FJ205416 ACR15984.1 KP690084 ALO61086.1 ODYU01001143 SOQ36937.1 PCG78736.1 RSAL01000093 RVE47909.1 FJ205411 ACR15979.2 JQ429439 AFW03964.1 KP690083 ALO61085.1 ODYU01007484 SOQ50292.1 ODYU01004626 SOQ44697.1 PCG78737.1 AY618894 AAV33657.1 EU673453 ACI45416.1 HM990180 ADT80828.1 EU673452 ACI45415.1 Y12272 CAA72951.1 GDQN01001948 JAT89106.1 KM360194 AKH49610.1 GDQN01005223 JAT85831.1 RVE47905.1 NWSH01000207 PCG78410.1 PCG78414.1 JN252040 AFM28253.1 PCG78412.1 AB026735 BAA77401.1 PCG78411.1 PZC77162.1 EF600058 ABU98623.1 PCG78413.1 MH201547 QAR48640.1 BABH01004141 BABH01004139 BABH01004140 PCG78415.1 KPI99023.1 PZC77163.1 PCG78408.1 FJ205435 ACR16001.2 AGBW02007651 OWR55096.1 ODYU01005353 SOQ46109.1 AGBW02009787 OWR49920.1 FJ205398 ACR15967.2 OWR55112.1 GDQN01010016 JAT81038.1 KPI98932.1 KQ459700 KPJ20347.1 ODYU01003737 SOQ42892.1 BABH01004171 BABH01004172 RSAL01000071 RVE49104.1 PCG78407.1 AY829820 AAV91434.2 ODYU01004804 SOQ45062.1 NWSH01000261 PCG77915.1 ODYU01005031 SOQ45511.1 ATLV01018958 KE525256 KFB43479.1 KX951431 ARR75599.1 JN252039 AFM28252.1 CVRI01000022 CRK92071.1 AY155562 AAN75000.1 GANO01002186 JAB57685.1 JXUM01007351 KQ560238 KXJ83665.1 GEZM01046011 JAV77579.1 JXUM01007352 KXJ83666.1 DS231893 EDS44181.1 AY155564 AAN75002.1 GFDL01005094 JAV29951.1 GFDL01004648 JAV30397.1 GFDL01004647 JAV30398.1 AY155563 AAN75001.1 CH478002 EAT34483.1 AF268665 AAG10005.1 AY603563 AAU06478.1 EDS44178.1 AY198134 AAO43403.1 AY995188 AAX94742.1 FX985331 BAX07344.1 EDS44176.1 KQ971348 EFA05747.1 KPI99004.1 M77814 AAA29356.1 AXCM01001005 EAT34478.1 AF266757 AF187078 AAF82286.1 AAG17021.1
Proteomes
Interpro
CDD
ProteinModelPortal
H9JTC9
A0A194Q127
A0A2W1BS57
A0A2A4K4P0
Q9BMQ7
J7FDE1
+ More
A0A0N1IP15 A0A0K8S2W5 A0A2W1BWI0 B4X992 H9JTD0 C9W8H1 A0A0X9I156 A0A2H1V7Z9 A0A2A4K3Y4 A0A437BBW9 C9W8G6 T1QDG4 A0A109P6B5 A0A2H1WBT2 A0A2H1VV25 A0A2A4K3B0 A0PGD9 B6D1R0 E7D010 B6D1Q9 O18437 A0A1E1WQI5 A0A1B0RHN9 A0A1E1WG34 A0A3S2LZV4 A0A2A4K409 A0A2A4K3D4 J7FDV5 A0A2A4K426 Q9XY10 A0A2A4K2C1 A0A2W1BQ29 B1NLE3 A0A2A4K3S1 A0A410KK64 H9JTC5 H9JTC6 A0A2A4K2C9 A0A194Q123 A0A2W1BZC6 A0A2A4K3B1 C9W8I8 A0A212FMW9 A0A2H1VZ50 A0A212F859 C9W8F4 A0A212FMY7 A0A1E1W214 A0A194Q024 A0A194RS93 A0A2H1VQY4 H9JTB5 A0A3S2NJF1 A0A2A4K2D7 Q5MGG6 A0A2H1VX31 A0A2A4K0W2 A0A2H1VXD9 A0A182IVJ9 A0A084VZT5 A0A2Z2GQC5 J7FCI3 A0A1J1HVG8 Q8I8E4 U5EJM0 A0A182G5U5 A0A1Y1LVG1 A0A182G5U6 B0WD46 Q8I8E2 A0A182K8Q1 A0A1Q3FQQ0 A0A1Q3FS52 A0A1Q3FS75 A0A1S4FZ18 Q8I8E3 Q16JM8 Q9GTK7 Q66UD0 B0WD43 A0A1I8NFS9 Q86PL8 A0A182SGP5 Q52NW5 A0A1V1FIT7 B0WD41 D2A3W9 A0A194Q1Y2 Q27440 A0A182MAA4 Q16JN3 Q9NB49
A0A0N1IP15 A0A0K8S2W5 A0A2W1BWI0 B4X992 H9JTD0 C9W8H1 A0A0X9I156 A0A2H1V7Z9 A0A2A4K3Y4 A0A437BBW9 C9W8G6 T1QDG4 A0A109P6B5 A0A2H1WBT2 A0A2H1VV25 A0A2A4K3B0 A0PGD9 B6D1R0 E7D010 B6D1Q9 O18437 A0A1E1WQI5 A0A1B0RHN9 A0A1E1WG34 A0A3S2LZV4 A0A2A4K409 A0A2A4K3D4 J7FDV5 A0A2A4K426 Q9XY10 A0A2A4K2C1 A0A2W1BQ29 B1NLE3 A0A2A4K3S1 A0A410KK64 H9JTC5 H9JTC6 A0A2A4K2C9 A0A194Q123 A0A2W1BZC6 A0A2A4K3B1 C9W8I8 A0A212FMW9 A0A2H1VZ50 A0A212F859 C9W8F4 A0A212FMY7 A0A1E1W214 A0A194Q024 A0A194RS93 A0A2H1VQY4 H9JTB5 A0A3S2NJF1 A0A2A4K2D7 Q5MGG6 A0A2H1VX31 A0A2A4K0W2 A0A2H1VXD9 A0A182IVJ9 A0A084VZT5 A0A2Z2GQC5 J7FCI3 A0A1J1HVG8 Q8I8E4 U5EJM0 A0A182G5U5 A0A1Y1LVG1 A0A182G5U6 B0WD46 Q8I8E2 A0A182K8Q1 A0A1Q3FQQ0 A0A1Q3FS52 A0A1Q3FS75 A0A1S4FZ18 Q8I8E3 Q16JM8 Q9GTK7 Q66UD0 B0WD43 A0A1I8NFS9 Q86PL8 A0A182SGP5 Q52NW5 A0A1V1FIT7 B0WD41 D2A3W9 A0A194Q1Y2 Q27440 A0A182MAA4 Q16JN3 Q9NB49
PDB
2HLC
E-value=9.86793e-21,
Score=245
Ontologies
GO
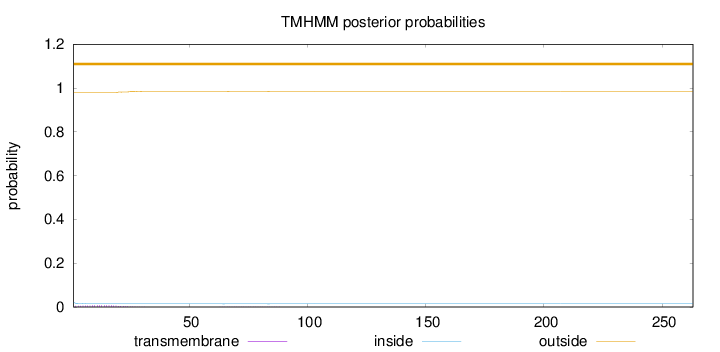
Topology
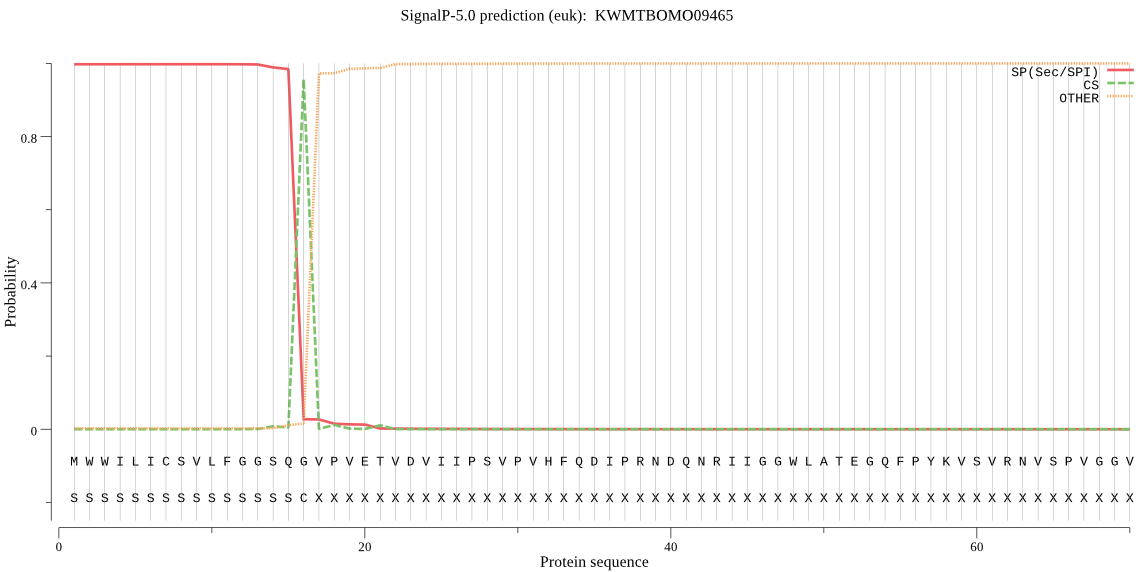
SignalP
Position: 1 - 16,
Likelihood: 0.997362
Length:
263
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.158310000000001
Exp number, first 60 AAs:
0.14389
Total prob of N-in:
0.02158
outside
1 - 263
Population Genetic Test Statistics
Pi
316.850452
Theta
175.352885
Tajima's D
2.756788
CLR
0.170223
CSRT
0.967451627418629
Interpretation
Uncertain
