Gene
KWMTBOMO09464
Pre Gene Modal
BGIBMGA012790
Annotation
PREDICTED:_prestin_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.846
Sequence
CDS
ATGCGTATAAAAAAGACGTTTCTGGTTTACGAAAATGAAAACAAATGCCGCAGTGAGAAGGAGCCTTTACTGCAAGATTCGGATAACAGTGAGGATTATTGGCTCAGCTCTCGTCCCAGTCAGCAGTTCAACGTCATCAGACGGGTCTACCAGCAAGATGCGCTCAACAAGGACGCCGATTACCAAGTTCCTGATAGACCACTGGGTCACCGCTGCAAAAAACTGGTCAACAGTTGCGGCATCGGTGAATGTCTCCTCAATTCAGTGCCGCTGATCAGATGGCTGCCGAAATACAAAACTAAAGAGTATCTCGTCGGGGATTTGGTTGCTGGAGCTACCACCGCTGTTATGCACATACCTCAAGGTATGGCGTACGCGTTGCTGTCAGAAGTACCACCGATAGTTGGATTGTATATGGCATTTTTCCCGGTGCTAATCTACGTTATATTCGGTACCTCCCCTCATATTTCCATGGGAACATTCGCGGTCGCTTGCTTGATGGCTGGAAAGGTTGTGACACAGCACGCTACGGTTCTAGACGTGACCCATGGGAACAGCACGAATGGTAGTACAGCGCCTGAAACTGGTATAGAATACACACCCATACAAGTTGTAAGCGTCGTTTGCCTCTGCGTTGGCATGTATCAAATCGTGATGTGGATGTTTCGTCTCGGTGCAGTATCCACTTTGCTGTCAGATTCCTTGGTGTCAGGATTCACGACGGCCGCCTCATTCCAAGTCGTCGCCTCTCAGCTGAAGGACCTGTTTGGGGTCAAGATAGGGAAGACGTCCGGGAACTATAAAGTCGTCTTTACAATAGTTGAGGTGGTGAAAAATCTTCCGAACCTGAACTGGGCTGCGTTCGTGATATCGCTTATCACTTGCATCGTCATAGCTTTGAACAATGAACTGCTTAAGCCGTGGGTGAGTAAACGCAGTAGAGTGCCAGTTCCTGTAGAGCTATTGGCCATCGTCGTCGGCACGTTGGTCTCCAAGTTCGGGGGGCTGAAGGAGCAATTCGGCATCAGTTTGGTCGGACATATACCGACCGGCCTTCCGGTGCCGCAAGTACCCGCCATCGAGCTGATCCCGCAAGTGGCCGTGGACGCCTTCACCATAACGATGGTGACCTATACCATATCGATGTCCATGGCTTTAATATTCGCGGCCAAGGAGAAATACGAGATCGACGCCAATCAAGAATTATTAGCGCTTGGAGCCAGTAACGTGTTCGCTTCGTTCTTCTGCTGCATACCTTTCTGTGCCAGTCTGTCGCGCTCGTACATACAGTACCAGGCGGGTAGCAAGACGGGAGTGACGTCAGTCGTGAGTGCGTCCCTCATCCTCTGCGTGCTGCTGTGGATCGGGCCATTCTTCGAGGACCTCCCTCGGTGCGTGCTGGCCGGCATAATCGTGGTCTCGCTCAAGGGAATGTTCATGCAGATGAAGGAGCTTACGAAATTCTGGGGTCTCAGTAAGCTGGACGCCATGGTGTGGCTGATCACTTTCCTGGTGACGCTAATCGTGAACATCGATATCGGTCTCGGGGCGGGTCTGCTGGCCAGCGTGGGCGCCTTGTTCTGCCGCTCCCAGAAACCCTACACCTGTCTCCTGGGCAGAGTTCTGGACACCGATCTCTACCTTGACACGAAGAGATACAGAGCAGCAGAAGAGCTGCCGGGCATCAAGATATTCCACTACTGCGGCGGCTTGAACTTCGCCAGCAAGAACCTGTTCCGGTCGACTCTCTTCCGGAAGATCGGGTACATGAAACCGGCCGTCGTGACGCACGACGAGGACAACAACCTCACCAAGAGCGAATCCTACGAATGGGATCCCAGTATCAGTCACAAGGTACAATGTGTGATAATAGACGCAACCGCTTTGTCGTACGTTGACGCGCCCGGGATCAAGAGTTTGATAGCAGTTCAGAGAGAACTAGTTAGCAGCAATATTACGGTGCTATTGGCGGGAGCTAACGGGCCCGTGCTGGAAATGATCGAGAGATACAACAATTTGGAGGCCGAGAGACTTCAGTTGGACACGTTCCCGACCGTGCACGACGCGGTCGTTTACTACAACGTATCAGAGGCAAGAAAGCACGACGTACCCGTCACGATAGCGCCGTGA
Protein
MRIKKTFLVYENENKCRSEKEPLLQDSDNSEDYWLSSRPSQQFNVIRRVYQQDALNKDADYQVPDRPLGHRCKKLVNSCGIGECLLNSVPLIRWLPKYKTKEYLVGDLVAGATTAVMHIPQGMAYALLSEVPPIVGLYMAFFPVLIYVIFGTSPHISMGTFAVACLMAGKVVTQHATVLDVTHGNSTNGSTAPETGIEYTPIQVVSVVCLCVGMYQIVMWMFRLGAVSTLLSDSLVSGFTTAASFQVVASQLKDLFGVKIGKTSGNYKVVFTIVEVVKNLPNLNWAAFVISLITCIVIALNNELLKPWVSKRSRVPVPVELLAIVVGTLVSKFGGLKEQFGISLVGHIPTGLPVPQVPAIELIPQVAVDAFTITMVTYTISMSMALIFAAKEKYEIDANQELLALGASNVFASFFCCIPFCASLSRSYIQYQAGSKTGVTSVVSASLILCVLLWIGPFFEDLPRCVLAGIIVVSLKGMFMQMKELTKFWGLSKLDAMVWLITFLVTLIVNIDIGLGAGLLASVGALFCRSQKPYTCLLGRVLDTDLYLDTKRYRAAEELPGIKIFHYCGGLNFASKNLFRSTLFRKIGYMKPAVVTHDEDNNLTKSESYEWDPSISHKVQCVIIDATALSYVDAPGIKSLIAVQRELVSSNITVLLAGANGPVLEMIERYNNLEAERLQLDTFPTVHDAVVYYNVSEARKHDVPVTIAP
Summary
Similarity
Belongs to the SLC26A/SulP transporter (TC 2.A.53) family.
Uniprot
H9JTC8
A0A2W1BVH2
A0A194Q0C0
A0A3S2PD14
A0A2A4K3T7
A0A212FMY5
+ More
A0A2H1WA54 E2AK49 A0A1W4WHS8 A0A0J7KPG9 A0A1W4WTN4 A0A1W4WTZ1 A0A154P4D0 A0A151JYY3 D2A5I3 A0A195BGJ1 A0A1Y1LH55 A0A151X577 F4WS23 A0A158NJF0 A0A3L8DJ00 A0A151J2C5 A0A026W734 A0A0L7QR03 A0A1Y1LEY7 A0A336K2Y3 A0A2A3EDK1 A0A195CER1 K7J754 A0A336LI86 A0A2M4BFP0 A0A2M4BFV0 A0A336MP87 D6WD12 A0A139WNP1 A0A2J7RMZ4 A0A232FF36 V5GTW3 A0A1Q3EVH3 A0A2M3Z5I7 A0A2M3Z5P6 A0A1Q3EVM3 A0A2M3Z5J4 A0A2J7RMY1 A0A1Q3EVG4 N6TLZ6 U5ETJ5 A0A182FTF5 A0A0N0U7C0 A0A023EVP7 A0A1W4XG85 A0A232EUL3 A0A182NCB2 A0A084V9Z0 A0A182GZ34 A0A182QUW9 A0A182J9Y6 A0A182PRM0 A0A182I5X7 A0A453YZP6 E3SB72 A0A182KV43 A0A182WXV6 A0A026WFU1 A0A182UVE6 A0A195CK24 A0A088ASI9 A0A088A5D9 A0A1Y1K1F3 E2AAM0 A0A0L7R858 A0A151X5G6 A0A151I139 A0A195EVM6 A0A154NZ29 F4X227 A0A2M4AMK0 U4U307 A0A151J0H9 W5JSX4 A0A182Y6S7 E2B6X7 A0A1L8DEE2 A0A1L8DEN5 E9I9M5 A0A182RL36 A0A2A3E6J2 A0A0M9A5H9 A0A182M3G9 A0A1B6KMM7 A0A1L8DEK3 A0A1Q3F0C8 A0A1Q3F0D2 A0A1Q3F048 Q17IP1 B0WRI0 Q5TUJ1 A0A1B0CWQ8 K7IWY8
A0A2H1WA54 E2AK49 A0A1W4WHS8 A0A0J7KPG9 A0A1W4WTN4 A0A1W4WTZ1 A0A154P4D0 A0A151JYY3 D2A5I3 A0A195BGJ1 A0A1Y1LH55 A0A151X577 F4WS23 A0A158NJF0 A0A3L8DJ00 A0A151J2C5 A0A026W734 A0A0L7QR03 A0A1Y1LEY7 A0A336K2Y3 A0A2A3EDK1 A0A195CER1 K7J754 A0A336LI86 A0A2M4BFP0 A0A2M4BFV0 A0A336MP87 D6WD12 A0A139WNP1 A0A2J7RMZ4 A0A232FF36 V5GTW3 A0A1Q3EVH3 A0A2M3Z5I7 A0A2M3Z5P6 A0A1Q3EVM3 A0A2M3Z5J4 A0A2J7RMY1 A0A1Q3EVG4 N6TLZ6 U5ETJ5 A0A182FTF5 A0A0N0U7C0 A0A023EVP7 A0A1W4XG85 A0A232EUL3 A0A182NCB2 A0A084V9Z0 A0A182GZ34 A0A182QUW9 A0A182J9Y6 A0A182PRM0 A0A182I5X7 A0A453YZP6 E3SB72 A0A182KV43 A0A182WXV6 A0A026WFU1 A0A182UVE6 A0A195CK24 A0A088ASI9 A0A088A5D9 A0A1Y1K1F3 E2AAM0 A0A0L7R858 A0A151X5G6 A0A151I139 A0A195EVM6 A0A154NZ29 F4X227 A0A2M4AMK0 U4U307 A0A151J0H9 W5JSX4 A0A182Y6S7 E2B6X7 A0A1L8DEE2 A0A1L8DEN5 E9I9M5 A0A182RL36 A0A2A3E6J2 A0A0M9A5H9 A0A182M3G9 A0A1B6KMM7 A0A1L8DEK3 A0A1Q3F0C8 A0A1Q3F0D2 A0A1Q3F048 Q17IP1 B0WRI0 Q5TUJ1 A0A1B0CWQ8 K7IWY8
Pubmed
EMBL
BABH01004124
BABH01004125
BABH01004126
BABH01004127
KZ149935
PZC77197.1
+ More
KQ459582 KPI99027.1 RSAL01000093 RVE47908.1 NWSH01000176 PCG78719.1 AGBW02007651 OWR55098.1 ODYU01007294 SOQ49948.1 GL440191 EFN66192.1 LBMM01004605 KMQ92247.1 KQ434803 KZC05980.1 KQ981440 KYN41662.1 KQ971345 EFA05376.1 KQ976480 KYM83757.1 GEZM01057611 JAV72221.1 KQ982543 KYQ55388.1 GL888295 EGI63002.1 ADTU01017666 QOIP01000007 RLU20420.1 KQ980414 KYN16146.1 KK107364 EZA51917.1 KQ414785 KOC60989.1 GEZM01057612 JAV72219.1 UFQS01000010 UFQT01000010 SSW97133.1 SSX17519.1 KZ288271 PBC29788.1 KQ977935 KYM98553.1 AAZX01008222 AAZX01015634 SSW97134.1 SSX17520.1 GGFJ01002711 MBW51852.1 GGFJ01002712 MBW51853.1 UFQT01001519 SSX30899.1 KQ971311 EEZ98816.1 KYB29497.1 NEVH01002547 PNF42192.1 NNAY01000297 OXU29376.1 GALX01004788 JAB63678.1 GFDL01015769 JAV19276.1 GGFM01003029 MBW23780.1 GGFM01003073 MBW23824.1 GFDL01015777 JAV19268.1 GGFM01003035 MBW23786.1 PNF42194.1 GFDL01015779 JAV19266.1 APGK01019138 APGK01019139 KB740092 KB632429 ENN81469.1 ERL95424.1 GANO01002752 JAB57119.1 KQ435710 KOX79932.1 GAPW01000340 JAC13258.1 NNAY01002117 OXU22031.1 ATLV01000833 KE523910 KFB34784.1 JXUM01021464 JXUM01021465 JXUM01021466 KQ560601 KXJ81685.1 AXCN02001765 APCN01003536 APCN01003537 AAAB01008848 GQ332421 ADE05544.1 KK107231 QOIP01000009 EZA54985.1 RLU19096.1 KQ977642 KYN01063.1 GEZM01097099 JAV54321.1 GL438129 EFN69527.1 KQ414633 KOC67065.1 KQ982519 KYQ55510.1 KQ976600 KYM79457.1 KQ981954 KYN32196.1 KQ434783 KZC04929.1 GL888565 EGI59443.1 GGFK01008683 MBW42004.1 KB631646 ERL84981.1 KQ980629 KYN15016.1 ADMH02000540 ETN66025.1 GL446060 EFN88552.1 GFDF01009273 JAV04811.1 GFDF01009274 JAV04810.1 GL761846 EFZ22685.1 KZ288364 PBC26892.1 KQ435729 KOX77640.1 AXCM01001425 GEBQ01027290 JAT12687.1 GFDF01009277 JAV04807.1 GFDL01014035 JAV21010.1 GFDL01014032 JAV21013.1 GFDL01014115 JAV20930.1 CH477238 EAT46519.1 DS232056 EDS33378.1 EAL41086.3 AJWK01032772 AAZX01010099 AAZX01012980
KQ459582 KPI99027.1 RSAL01000093 RVE47908.1 NWSH01000176 PCG78719.1 AGBW02007651 OWR55098.1 ODYU01007294 SOQ49948.1 GL440191 EFN66192.1 LBMM01004605 KMQ92247.1 KQ434803 KZC05980.1 KQ981440 KYN41662.1 KQ971345 EFA05376.1 KQ976480 KYM83757.1 GEZM01057611 JAV72221.1 KQ982543 KYQ55388.1 GL888295 EGI63002.1 ADTU01017666 QOIP01000007 RLU20420.1 KQ980414 KYN16146.1 KK107364 EZA51917.1 KQ414785 KOC60989.1 GEZM01057612 JAV72219.1 UFQS01000010 UFQT01000010 SSW97133.1 SSX17519.1 KZ288271 PBC29788.1 KQ977935 KYM98553.1 AAZX01008222 AAZX01015634 SSW97134.1 SSX17520.1 GGFJ01002711 MBW51852.1 GGFJ01002712 MBW51853.1 UFQT01001519 SSX30899.1 KQ971311 EEZ98816.1 KYB29497.1 NEVH01002547 PNF42192.1 NNAY01000297 OXU29376.1 GALX01004788 JAB63678.1 GFDL01015769 JAV19276.1 GGFM01003029 MBW23780.1 GGFM01003073 MBW23824.1 GFDL01015777 JAV19268.1 GGFM01003035 MBW23786.1 PNF42194.1 GFDL01015779 JAV19266.1 APGK01019138 APGK01019139 KB740092 KB632429 ENN81469.1 ERL95424.1 GANO01002752 JAB57119.1 KQ435710 KOX79932.1 GAPW01000340 JAC13258.1 NNAY01002117 OXU22031.1 ATLV01000833 KE523910 KFB34784.1 JXUM01021464 JXUM01021465 JXUM01021466 KQ560601 KXJ81685.1 AXCN02001765 APCN01003536 APCN01003537 AAAB01008848 GQ332421 ADE05544.1 KK107231 QOIP01000009 EZA54985.1 RLU19096.1 KQ977642 KYN01063.1 GEZM01097099 JAV54321.1 GL438129 EFN69527.1 KQ414633 KOC67065.1 KQ982519 KYQ55510.1 KQ976600 KYM79457.1 KQ981954 KYN32196.1 KQ434783 KZC04929.1 GL888565 EGI59443.1 GGFK01008683 MBW42004.1 KB631646 ERL84981.1 KQ980629 KYN15016.1 ADMH02000540 ETN66025.1 GL446060 EFN88552.1 GFDF01009273 JAV04811.1 GFDF01009274 JAV04810.1 GL761846 EFZ22685.1 KZ288364 PBC26892.1 KQ435729 KOX77640.1 AXCM01001425 GEBQ01027290 JAT12687.1 GFDF01009277 JAV04807.1 GFDL01014035 JAV21010.1 GFDL01014032 JAV21013.1 GFDL01014115 JAV20930.1 CH477238 EAT46519.1 DS232056 EDS33378.1 EAL41086.3 AJWK01032772 AAZX01010099 AAZX01012980
Proteomes
UP000005204
UP000053268
UP000283053
UP000218220
UP000007151
UP000000311
+ More
UP000192223 UP000036403 UP000076502 UP000078541 UP000007266 UP000078540 UP000075809 UP000007755 UP000005205 UP000279307 UP000078492 UP000053097 UP000053825 UP000242457 UP000078542 UP000002358 UP000235965 UP000215335 UP000019118 UP000030742 UP000069272 UP000053105 UP000075884 UP000030765 UP000069940 UP000249989 UP000075886 UP000075880 UP000075885 UP000075840 UP000075882 UP000076407 UP000075903 UP000005203 UP000000673 UP000076408 UP000008237 UP000075900 UP000075883 UP000008820 UP000002320 UP000007062 UP000092461
UP000192223 UP000036403 UP000076502 UP000078541 UP000007266 UP000078540 UP000075809 UP000007755 UP000005205 UP000279307 UP000078492 UP000053097 UP000053825 UP000242457 UP000078542 UP000002358 UP000235965 UP000215335 UP000019118 UP000030742 UP000069272 UP000053105 UP000075884 UP000030765 UP000069940 UP000249989 UP000075886 UP000075880 UP000075885 UP000075840 UP000075882 UP000076407 UP000075903 UP000005203 UP000000673 UP000076408 UP000008237 UP000075900 UP000075883 UP000008820 UP000002320 UP000007062 UP000092461
PRIDE
Interpro
SUPFAM
SSF52091
SSF52091
Gene 3D
ProteinModelPortal
H9JTC8
A0A2W1BVH2
A0A194Q0C0
A0A3S2PD14
A0A2A4K3T7
A0A212FMY5
+ More
A0A2H1WA54 E2AK49 A0A1W4WHS8 A0A0J7KPG9 A0A1W4WTN4 A0A1W4WTZ1 A0A154P4D0 A0A151JYY3 D2A5I3 A0A195BGJ1 A0A1Y1LH55 A0A151X577 F4WS23 A0A158NJF0 A0A3L8DJ00 A0A151J2C5 A0A026W734 A0A0L7QR03 A0A1Y1LEY7 A0A336K2Y3 A0A2A3EDK1 A0A195CER1 K7J754 A0A336LI86 A0A2M4BFP0 A0A2M4BFV0 A0A336MP87 D6WD12 A0A139WNP1 A0A2J7RMZ4 A0A232FF36 V5GTW3 A0A1Q3EVH3 A0A2M3Z5I7 A0A2M3Z5P6 A0A1Q3EVM3 A0A2M3Z5J4 A0A2J7RMY1 A0A1Q3EVG4 N6TLZ6 U5ETJ5 A0A182FTF5 A0A0N0U7C0 A0A023EVP7 A0A1W4XG85 A0A232EUL3 A0A182NCB2 A0A084V9Z0 A0A182GZ34 A0A182QUW9 A0A182J9Y6 A0A182PRM0 A0A182I5X7 A0A453YZP6 E3SB72 A0A182KV43 A0A182WXV6 A0A026WFU1 A0A182UVE6 A0A195CK24 A0A088ASI9 A0A088A5D9 A0A1Y1K1F3 E2AAM0 A0A0L7R858 A0A151X5G6 A0A151I139 A0A195EVM6 A0A154NZ29 F4X227 A0A2M4AMK0 U4U307 A0A151J0H9 W5JSX4 A0A182Y6S7 E2B6X7 A0A1L8DEE2 A0A1L8DEN5 E9I9M5 A0A182RL36 A0A2A3E6J2 A0A0M9A5H9 A0A182M3G9 A0A1B6KMM7 A0A1L8DEK3 A0A1Q3F0C8 A0A1Q3F0D2 A0A1Q3F048 Q17IP1 B0WRI0 Q5TUJ1 A0A1B0CWQ8 K7IWY8
A0A2H1WA54 E2AK49 A0A1W4WHS8 A0A0J7KPG9 A0A1W4WTN4 A0A1W4WTZ1 A0A154P4D0 A0A151JYY3 D2A5I3 A0A195BGJ1 A0A1Y1LH55 A0A151X577 F4WS23 A0A158NJF0 A0A3L8DJ00 A0A151J2C5 A0A026W734 A0A0L7QR03 A0A1Y1LEY7 A0A336K2Y3 A0A2A3EDK1 A0A195CER1 K7J754 A0A336LI86 A0A2M4BFP0 A0A2M4BFV0 A0A336MP87 D6WD12 A0A139WNP1 A0A2J7RMZ4 A0A232FF36 V5GTW3 A0A1Q3EVH3 A0A2M3Z5I7 A0A2M3Z5P6 A0A1Q3EVM3 A0A2M3Z5J4 A0A2J7RMY1 A0A1Q3EVG4 N6TLZ6 U5ETJ5 A0A182FTF5 A0A0N0U7C0 A0A023EVP7 A0A1W4XG85 A0A232EUL3 A0A182NCB2 A0A084V9Z0 A0A182GZ34 A0A182QUW9 A0A182J9Y6 A0A182PRM0 A0A182I5X7 A0A453YZP6 E3SB72 A0A182KV43 A0A182WXV6 A0A026WFU1 A0A182UVE6 A0A195CK24 A0A088ASI9 A0A088A5D9 A0A1Y1K1F3 E2AAM0 A0A0L7R858 A0A151X5G6 A0A151I139 A0A195EVM6 A0A154NZ29 F4X227 A0A2M4AMK0 U4U307 A0A151J0H9 W5JSX4 A0A182Y6S7 E2B6X7 A0A1L8DEE2 A0A1L8DEN5 E9I9M5 A0A182RL36 A0A2A3E6J2 A0A0M9A5H9 A0A182M3G9 A0A1B6KMM7 A0A1L8DEK3 A0A1Q3F0C8 A0A1Q3F0D2 A0A1Q3F048 Q17IP1 B0WRI0 Q5TUJ1 A0A1B0CWQ8 K7IWY8
PDB
5DA0
E-value=5.98229e-15,
Score=199
Ontologies
GO
PANTHER
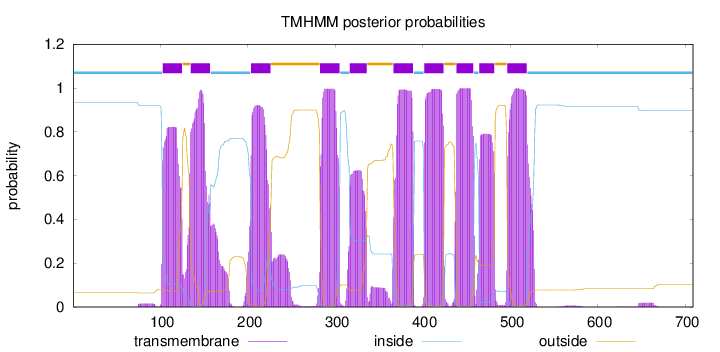
Topology
Subcellular location
Membrane
Length:
709
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
211.434030000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.93542
inside
1 - 102
TMhelix
103 - 125
outside
126 - 134
TMhelix
135 - 157
inside
158 - 203
TMhelix
204 - 226
outside
227 - 282
TMhelix
283 - 305
inside
306 - 316
TMhelix
317 - 336
outside
337 - 366
TMhelix
367 - 389
inside
390 - 401
TMhelix
402 - 424
outside
425 - 438
TMhelix
439 - 458
inside
459 - 464
TMhelix
465 - 482
outside
483 - 496
TMhelix
497 - 519
inside
520 - 709
Population Genetic Test Statistics
Pi
243.604142
Theta
189.144543
Tajima's D
1.099342
CLR
0.567847
CSRT
0.696065196740163
Interpretation
Uncertain
