Gene
KWMTBOMO09461
Pre Gene Modal
BGIBMGA012789
Annotation
PREDICTED:_ATP-binding_cassette_sub-family_A_member_5-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.944
Sequence
CDS
ATGCTGTGCGGTCTACTGCTGCTGTACAGTCCCTCCGCCATACTGTTCAATACGTGCCTCTCGTACATATTCGACAAGATGGACTCCGCCCAGAGCATCATGCCGAACATCACGACCTGGGTCGGAGTCATCCCGTTCATCTTGGTCGCTGTTCTCGACACCTTTAAATGGGGCAGCGACATCGCGTTCTACCTTCACCTGGTGTTCAGCTTCCTGGACGTCATGTACATACCGTACGCCATCATGTACTATGTCGACAGGGTATATGTGACGTGCAACCTCCGGGGGGTGTGCACTACCCCCACCCTGGGCAGCTACTTCACGGCGGAGGTGTGGGTGCTGATCGCCGCCATGCTGCTACACGTGCCGCTCTGCGGGGCCGCACTCCTCGCCGCCGACAAGCTCAAGACTGGGGGAATCGCGGCCTTTAGAAGTCGCAAGCGCTCCGAGACGAGAGCGGGGAGCGTGAGCGGCGCGGGGGTAGGGGCGGCGGGCGCGGGGGGCGAGGAGGCGGCGTACGGCACGGAGGGGGGCGAGGACGGTGACGTCAAGCGAGAGAGACGACGGGTCGCCAACATACTCGCCAGCAACCAGAACAACATGCCACCCTTACTTGTTCATAACCTTCGGAAAGAGTACAAAGTGCGCGGTACGGAGGACGGTGGGGGCTGCTTTGGTGACAGCAAGAGTCCCACGCGGCGGTCTCGGAGGACCGGCCTGGCGAGGCTGAGCTTGGCCGTGGAGGGAGGGGAGGTGTTCGGGCTCTTGGGCCATAACGGAGCCGGGAAAACCACTACAATGAAGATTATTACTGCCGAAGAGAGACCTACTTGTGGAACGGTAATGCTTGGTAGTCAAGACATAAGGGATTCCCAAGCGTCAGCGTTCCGCATGCTGGGCTACTGTCCGCAGCACGACGCGCTGTGGAAGAACGTCACCATCCGGGAGCACATCGAGTGCTACGCCGCCATCCGGGGGGTCAGCAACCAGGACATACCCCGGATCGTGGACGCATACCTAAACGGTCTTCAAATAGCAGAGCACGCCGGTAAGAACGCGGAGGAATGCTCCGGGGGAACTCGTCGTAAGCTGTCCTTTGCTTTGGCCATGGTGGGTGCACCACGCGTGGTGCTGCTAGACGAGCCCTCCACCGGGATGGACCCGAGGAGCAAGAGATTCCTATGGGACACCATTCTTGCCAGCTTCCAGGGTAAGAAAGGTGCCATACTAACGACACATTCCATGGAAGAGGCTGATGCATTATGTTCCAGAGTTGGGATCATGGTTAAGGGGAGTTTGAGGTGTATCGGTTCAACGCAACACTTGAAGAATCTATACGGAGCCGGGTACACGCTGGAGATGAAGATAGGGCAGACCAATCAGAAATCAACGGTATGTTAA
Protein
MLCGLLLLYSPSAILFNTCLSYIFDKMDSAQSIMPNITTWVGVIPFILVAVLDTFKWGSDIAFYLHLVFSFLDVMYIPYAIMYYVDRVYVTCNLRGVCTTPTLGSYFTAEVWVLIAAMLLHVPLCGAALLAADKLKTGGIAAFRSRKRSETRAGSVSGAGVGAAGAGGEEAAYGTEGGEDGDVKRERRRVANILASNQNNMPPLLVHNLRKEYKVRGTEDGGGCFGDSKSPTRRSRRTGLARLSLAVEGGEVFGLLGHNGAGKTTTMKIITAEERPTCGTVMLGSQDIRDSQASAFRMLGYCPQHDALWKNVTIREHIECYAAIRGVSNQDIPRIVDAYLNGLQIAEHAGKNAEECSGGTRRKLSFALAMVGAPRVVLLDEPSTGMDPRSKRFLWDTILASFQGKKGAILTTHSMEEADALCSRVGIMVKGSLRCIGSTQHLKNLYGAGYTLEMKIGQTNQKSTVC
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
H9JTC7
A0A2W1BQ17
A0A2A4IY78
A0A3S2LJA8
A0A2H1V1C0
A0A194Q201
+ More
E2ACV7 A0A0T6BE97 A0A2J7PMF3 A0A2J7PME9 A0A067R5U3 A0A026WAC0 A0A3L8DZH5 A0A154PKS0 A0A0C9QXZ8 A0A0C9R779 A0A2A3E528 A0A0L7R8K5 A0A088A6H6 E2BWV8 E0VHQ3 A0A0M8ZPX7 K7IRX5 A0A232EPW5 D2A4C6 A0A1Y1ME33 A0A182T3A9 A0A151JMM8 A0A195BY13 A0A182UU28 U4U0E3 A0A195CTJ9 A0A0J7KX87 A0A195F057 A0A2J7PME8 A0A182J101 A0A084VBW2 A0A1Y9HDT4 A0A182XB03 A0A182HX76 A0A1S4H3X5 A0A182PEV1 A0A182YLY5 A0A182W3R0 W5JB00 A0A182NDJ3 A0A182FM13 A0A182MP80 A0A182PZJ3 B0WRK5 A0A0N1IFN1 A0A0P5DKF7 A0A0P5CFB7 A0A0P5CMY9 A0A0P5KBG2 A0A151XGD7 A0A0P5HP52 A0A2P8ZKQ4 A0A0P5FVZ1 A0A0N8CGI5 A0A0P5SAB4 A0A0P5KW04 A0A0N8B135 A0A0P5V6X4 A0A0N8CLT5 A0A0P5CN38 A0A0N8A741 A0A0P5I5G2 A0A0P6GIR6 A0A0P6A7S8 A0A0P6G9I9 A0A0N8EK62 A0A0P6FNG6 A0A0P5QGE7 A0A0P6F0D9 A0A0P5PQL1 A0A0P5NZK9 A0A0P5LAR9 A0A0P5V5S4 A0A410GHQ8 E9FSC6 J9K233 A0A0P4XQ78 A0A0A7AS68 A0A0P6BYZ9 A0A158P427 Q17D43 A0A3R7PTA6 A0A0P5YQ47 A0A1B6CH43 A0A1B6DZ36 T1JC73 A0A1B6L4N1 A0A1B6L2P7 A0A1D2ML69
E2ACV7 A0A0T6BE97 A0A2J7PMF3 A0A2J7PME9 A0A067R5U3 A0A026WAC0 A0A3L8DZH5 A0A154PKS0 A0A0C9QXZ8 A0A0C9R779 A0A2A3E528 A0A0L7R8K5 A0A088A6H6 E2BWV8 E0VHQ3 A0A0M8ZPX7 K7IRX5 A0A232EPW5 D2A4C6 A0A1Y1ME33 A0A182T3A9 A0A151JMM8 A0A195BY13 A0A182UU28 U4U0E3 A0A195CTJ9 A0A0J7KX87 A0A195F057 A0A2J7PME8 A0A182J101 A0A084VBW2 A0A1Y9HDT4 A0A182XB03 A0A182HX76 A0A1S4H3X5 A0A182PEV1 A0A182YLY5 A0A182W3R0 W5JB00 A0A182NDJ3 A0A182FM13 A0A182MP80 A0A182PZJ3 B0WRK5 A0A0N1IFN1 A0A0P5DKF7 A0A0P5CFB7 A0A0P5CMY9 A0A0P5KBG2 A0A151XGD7 A0A0P5HP52 A0A2P8ZKQ4 A0A0P5FVZ1 A0A0N8CGI5 A0A0P5SAB4 A0A0P5KW04 A0A0N8B135 A0A0P5V6X4 A0A0N8CLT5 A0A0P5CN38 A0A0N8A741 A0A0P5I5G2 A0A0P6GIR6 A0A0P6A7S8 A0A0P6G9I9 A0A0N8EK62 A0A0P6FNG6 A0A0P5QGE7 A0A0P6F0D9 A0A0P5PQL1 A0A0P5NZK9 A0A0P5LAR9 A0A0P5V5S4 A0A410GHQ8 E9FSC6 J9K233 A0A0P4XQ78 A0A0A7AS68 A0A0P6BYZ9 A0A158P427 Q17D43 A0A3R7PTA6 A0A0P5YQ47 A0A1B6CH43 A0A1B6DZ36 T1JC73 A0A1B6L4N1 A0A1B6L2P7 A0A1D2ML69
Pubmed
EMBL
BABH01004135
BABH01004136
BABH01004137
BABH01004138
KZ149935
PZC77159.1
+ More
NWSH01005566 PCG64122.1 RSAL01000093 RVE47906.1 ODYU01000187 SOQ34556.1 KQ459582 KPI99024.1 GL438582 EFN68755.1 LJIG01001320 KRT85620.1 NEVH01024021 PNF17507.1 PNF17508.1 KK852677 KDR18677.1 KK107323 EZA52591.1 QOIP01000002 RLU25880.1 KQ434936 KZC11898.1 GBYB01008659 JAG78426.1 GBYB01008657 JAG78424.1 KZ288405 PBC26181.1 KQ414632 KOC67217.1 GL451188 EFN79822.1 DS235171 EEB12939.1 KQ435922 KOX68527.1 NNAY01002890 OXU20378.1 KQ971348 EFA05649.1 GEZM01033856 JAV84079.1 KQ978901 KYN27579.1 KQ976394 KYM93185.1 KB631749 ERL85768.1 KQ977304 KYN03817.1 LBMM01002389 KMQ94889.1 KQ981880 KYN33955.1 PNF17510.1 ATLV01009630 KE524540 KFB35456.1 APCN01005148 AAAB01008849 ADMH02002024 ETN59939.1 AXCM01001041 AXCN02001759 DS232056 EDS33403.1 KQ460651 KPJ13052.1 GDIP01171909 JAJ51493.1 GDIP01171910 JAJ51492.1 GDIP01171908 JAJ51494.1 GDIQ01188036 JAK63689.1 KQ982174 KYQ59358.1 GDIQ01224474 JAK27251.1 PYGN01000027 PSN57079.1 GDIQ01249576 JAK02149.1 GDIP01133967 JAL69747.1 GDIP01142369 JAL61345.1 GDIQ01186440 JAK65285.1 GDIQ01223179 JAK28546.1 GDIP01103969 JAL99745.1 GDIP01167413 GDIP01149519 GDIP01119167 LRGB01003123 JAL84547.1 KZS04262.1 GDIP01167864 JAJ55538.1 GDIP01176009 JAJ47393.1 GDIQ01223180 JAK28545.1 GDIQ01035049 JAN59688.1 GDIP01038133 JAM65582.1 GDIQ01036651 JAN58086.1 GDIQ01018772 JAN75965.1 GDIQ01056735 JAN38002.1 GDIQ01135660 JAL16066.1 GDIQ01056736 JAN38001.1 GDIQ01135661 JAL16065.1 GDIQ01135662 JAL16064.1 GDIQ01172590 JAK79135.1 GDIP01103968 JAL99746.1 MH172493 QAA95900.1 GL732524 EFX89859.1 ABLF02039796 ABLF02039804 ABLF02039828 GDIP01238308 JAI85093.1 KF906265 AHK05630.1 GDIP01012164 JAM91551.1 ADTU01008287 ADTU01008288 ADTU01008289 ADTU01008290 CH477300 EAT44278.1 QCYY01000894 ROT82022.1 GDIP01054778 JAM48937.1 GEDC01024472 JAS12826.1 GEDC01006379 JAS30919.1 JH432062 GEBQ01021298 JAT18679.1 GEBQ01022051 JAT17926.1 LJIJ01000927 ODM93723.1
NWSH01005566 PCG64122.1 RSAL01000093 RVE47906.1 ODYU01000187 SOQ34556.1 KQ459582 KPI99024.1 GL438582 EFN68755.1 LJIG01001320 KRT85620.1 NEVH01024021 PNF17507.1 PNF17508.1 KK852677 KDR18677.1 KK107323 EZA52591.1 QOIP01000002 RLU25880.1 KQ434936 KZC11898.1 GBYB01008659 JAG78426.1 GBYB01008657 JAG78424.1 KZ288405 PBC26181.1 KQ414632 KOC67217.1 GL451188 EFN79822.1 DS235171 EEB12939.1 KQ435922 KOX68527.1 NNAY01002890 OXU20378.1 KQ971348 EFA05649.1 GEZM01033856 JAV84079.1 KQ978901 KYN27579.1 KQ976394 KYM93185.1 KB631749 ERL85768.1 KQ977304 KYN03817.1 LBMM01002389 KMQ94889.1 KQ981880 KYN33955.1 PNF17510.1 ATLV01009630 KE524540 KFB35456.1 APCN01005148 AAAB01008849 ADMH02002024 ETN59939.1 AXCM01001041 AXCN02001759 DS232056 EDS33403.1 KQ460651 KPJ13052.1 GDIP01171909 JAJ51493.1 GDIP01171910 JAJ51492.1 GDIP01171908 JAJ51494.1 GDIQ01188036 JAK63689.1 KQ982174 KYQ59358.1 GDIQ01224474 JAK27251.1 PYGN01000027 PSN57079.1 GDIQ01249576 JAK02149.1 GDIP01133967 JAL69747.1 GDIP01142369 JAL61345.1 GDIQ01186440 JAK65285.1 GDIQ01223179 JAK28546.1 GDIP01103969 JAL99745.1 GDIP01167413 GDIP01149519 GDIP01119167 LRGB01003123 JAL84547.1 KZS04262.1 GDIP01167864 JAJ55538.1 GDIP01176009 JAJ47393.1 GDIQ01223180 JAK28545.1 GDIQ01035049 JAN59688.1 GDIP01038133 JAM65582.1 GDIQ01036651 JAN58086.1 GDIQ01018772 JAN75965.1 GDIQ01056735 JAN38002.1 GDIQ01135660 JAL16066.1 GDIQ01056736 JAN38001.1 GDIQ01135661 JAL16065.1 GDIQ01135662 JAL16064.1 GDIQ01172590 JAK79135.1 GDIP01103968 JAL99746.1 MH172493 QAA95900.1 GL732524 EFX89859.1 ABLF02039796 ABLF02039804 ABLF02039828 GDIP01238308 JAI85093.1 KF906265 AHK05630.1 GDIP01012164 JAM91551.1 ADTU01008287 ADTU01008288 ADTU01008289 ADTU01008290 CH477300 EAT44278.1 QCYY01000894 ROT82022.1 GDIP01054778 JAM48937.1 GEDC01024472 JAS12826.1 GEDC01006379 JAS30919.1 JH432062 GEBQ01021298 JAT18679.1 GEBQ01022051 JAT17926.1 LJIJ01000927 ODM93723.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000000311
UP000235965
+ More
UP000027135 UP000053097 UP000279307 UP000076502 UP000242457 UP000053825 UP000005203 UP000008237 UP000009046 UP000053105 UP000002358 UP000215335 UP000007266 UP000075901 UP000078492 UP000078540 UP000075903 UP000030742 UP000078542 UP000036403 UP000078541 UP000075880 UP000030765 UP000075900 UP000076407 UP000075840 UP000075885 UP000076408 UP000075920 UP000000673 UP000075884 UP000069272 UP000075883 UP000075886 UP000002320 UP000053240 UP000075809 UP000245037 UP000076858 UP000000305 UP000007819 UP000005205 UP000008820 UP000283509 UP000094527
UP000027135 UP000053097 UP000279307 UP000076502 UP000242457 UP000053825 UP000005203 UP000008237 UP000009046 UP000053105 UP000002358 UP000215335 UP000007266 UP000075901 UP000078492 UP000078540 UP000075903 UP000030742 UP000078542 UP000036403 UP000078541 UP000075880 UP000030765 UP000075900 UP000076407 UP000075840 UP000075885 UP000076408 UP000075920 UP000000673 UP000075884 UP000069272 UP000075883 UP000075886 UP000002320 UP000053240 UP000075809 UP000245037 UP000076858 UP000000305 UP000007819 UP000005205 UP000008820 UP000283509 UP000094527
PRIDE
Interpro
IPR027417
P-loop_NTPase
+ More
IPR017871 ABC_transporter_CS
IPR003439 ABC_transporter-like
IPR003593 AAA+_ATPase
IPR020846 MFS_dom
IPR026082 ABCA
IPR011990 TPR-like_helical_dom_sf
IPR019412 Iml2/TPR_39
IPR019734 TPR_repeat
IPR001254 Trypsin_dom
IPR018114 TRYPSIN_HIS
IPR001314 Peptidase_S1A
IPR009003 Peptidase_S1_PA
IPR017871 ABC_transporter_CS
IPR003439 ABC_transporter-like
IPR003593 AAA+_ATPase
IPR020846 MFS_dom
IPR026082 ABCA
IPR011990 TPR-like_helical_dom_sf
IPR019412 Iml2/TPR_39
IPR019734 TPR_repeat
IPR001254 Trypsin_dom
IPR018114 TRYPSIN_HIS
IPR001314 Peptidase_S1A
IPR009003 Peptidase_S1_PA
Gene 3D
ProteinModelPortal
H9JTC7
A0A2W1BQ17
A0A2A4IY78
A0A3S2LJA8
A0A2H1V1C0
A0A194Q201
+ More
E2ACV7 A0A0T6BE97 A0A2J7PMF3 A0A2J7PME9 A0A067R5U3 A0A026WAC0 A0A3L8DZH5 A0A154PKS0 A0A0C9QXZ8 A0A0C9R779 A0A2A3E528 A0A0L7R8K5 A0A088A6H6 E2BWV8 E0VHQ3 A0A0M8ZPX7 K7IRX5 A0A232EPW5 D2A4C6 A0A1Y1ME33 A0A182T3A9 A0A151JMM8 A0A195BY13 A0A182UU28 U4U0E3 A0A195CTJ9 A0A0J7KX87 A0A195F057 A0A2J7PME8 A0A182J101 A0A084VBW2 A0A1Y9HDT4 A0A182XB03 A0A182HX76 A0A1S4H3X5 A0A182PEV1 A0A182YLY5 A0A182W3R0 W5JB00 A0A182NDJ3 A0A182FM13 A0A182MP80 A0A182PZJ3 B0WRK5 A0A0N1IFN1 A0A0P5DKF7 A0A0P5CFB7 A0A0P5CMY9 A0A0P5KBG2 A0A151XGD7 A0A0P5HP52 A0A2P8ZKQ4 A0A0P5FVZ1 A0A0N8CGI5 A0A0P5SAB4 A0A0P5KW04 A0A0N8B135 A0A0P5V6X4 A0A0N8CLT5 A0A0P5CN38 A0A0N8A741 A0A0P5I5G2 A0A0P6GIR6 A0A0P6A7S8 A0A0P6G9I9 A0A0N8EK62 A0A0P6FNG6 A0A0P5QGE7 A0A0P6F0D9 A0A0P5PQL1 A0A0P5NZK9 A0A0P5LAR9 A0A0P5V5S4 A0A410GHQ8 E9FSC6 J9K233 A0A0P4XQ78 A0A0A7AS68 A0A0P6BYZ9 A0A158P427 Q17D43 A0A3R7PTA6 A0A0P5YQ47 A0A1B6CH43 A0A1B6DZ36 T1JC73 A0A1B6L4N1 A0A1B6L2P7 A0A1D2ML69
E2ACV7 A0A0T6BE97 A0A2J7PMF3 A0A2J7PME9 A0A067R5U3 A0A026WAC0 A0A3L8DZH5 A0A154PKS0 A0A0C9QXZ8 A0A0C9R779 A0A2A3E528 A0A0L7R8K5 A0A088A6H6 E2BWV8 E0VHQ3 A0A0M8ZPX7 K7IRX5 A0A232EPW5 D2A4C6 A0A1Y1ME33 A0A182T3A9 A0A151JMM8 A0A195BY13 A0A182UU28 U4U0E3 A0A195CTJ9 A0A0J7KX87 A0A195F057 A0A2J7PME8 A0A182J101 A0A084VBW2 A0A1Y9HDT4 A0A182XB03 A0A182HX76 A0A1S4H3X5 A0A182PEV1 A0A182YLY5 A0A182W3R0 W5JB00 A0A182NDJ3 A0A182FM13 A0A182MP80 A0A182PZJ3 B0WRK5 A0A0N1IFN1 A0A0P5DKF7 A0A0P5CFB7 A0A0P5CMY9 A0A0P5KBG2 A0A151XGD7 A0A0P5HP52 A0A2P8ZKQ4 A0A0P5FVZ1 A0A0N8CGI5 A0A0P5SAB4 A0A0P5KW04 A0A0N8B135 A0A0P5V6X4 A0A0N8CLT5 A0A0P5CN38 A0A0N8A741 A0A0P5I5G2 A0A0P6GIR6 A0A0P6A7S8 A0A0P6G9I9 A0A0N8EK62 A0A0P6FNG6 A0A0P5QGE7 A0A0P6F0D9 A0A0P5PQL1 A0A0P5NZK9 A0A0P5LAR9 A0A0P5V5S4 A0A410GHQ8 E9FSC6 J9K233 A0A0P4XQ78 A0A0A7AS68 A0A0P6BYZ9 A0A158P427 Q17D43 A0A3R7PTA6 A0A0P5YQ47 A0A1B6CH43 A0A1B6DZ36 T1JC73 A0A1B6L4N1 A0A1B6L2P7 A0A1D2ML69
PDB
5XJY
E-value=5.3727e-57,
Score=560
Ontologies
GO
PANTHER
Topology
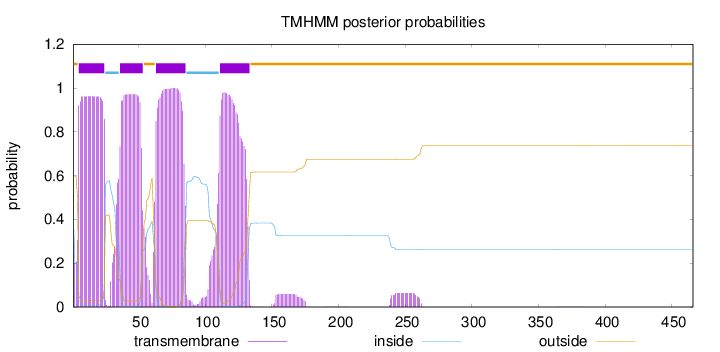
Length:
466
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
86.6952400000001
Exp number, first 60 AAs:
39.80544
Total prob of N-in:
0.40377
POSSIBLE N-term signal
sequence
outside
1 - 4
TMhelix
5 - 24
inside
25 - 35
TMhelix
36 - 53
outside
54 - 62
TMhelix
63 - 85
inside
86 - 110
TMhelix
111 - 133
outside
134 - 466
Population Genetic Test Statistics
Pi
186.34816
Theta
149.921963
Tajima's D
0.766174
CLR
0.031095
CSRT
0.592970351482426
Interpretation
Uncertain
