Gene
KWMTBOMO09460
Pre Gene Modal
BGIBMGA012789
Annotation
PREDICTED:_ATP-binding_cassette_sub-family_A_member_5-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.507
Sequence
CDS
ATGATCAGGGATCCCTACAAGCTGTACGTCATGATCTTCTTGCCTATAATATCCTGTGCCCTCGGTCTTTACATGAAGTCACGACAAATAGTCTTCTTCCGAATGCAGCCACTGACTTTGGGCCCAGATGCCTATTTCAACAAGACCCCCATAGCGCTGCATAGCGAACAGAATACTTTGAGAGAAGACTTCAAAGAATCGCTAGAGATCCTCGGAGCTCACCCTATAGTGGACTTCGATGGAAACTTCTCAAGTCTACTAGACATGGAGAACTTTGGGGCGTTTAGTCTGAAGGAGAGCTTAAATCCATATGGGAAGGTCATGGCGTATTATAACACAACTTACACGCACAGTTTGCCGATAATCATCAATTTGCTGGACAACAGTCTCTATAGGGCCCTAATGTCAGCATCGGGGCTGGAGCACCACTTCAAACCAATAGAAGTGATGGCCCATCCGTTCCAGCAGACAGAGCAGCAGGAAGAGTTCAATTTGGGCAACGTCATCTGCGCTGTGTTCGTCGGCATGATCTTCGCGCTGGTACCAGTGACCCTGGCTGTCGACATCGTCTACGACCGGGAGATCAAAGCAAAGAATCAACTTCGCGTCAACGGCCTCTCGATGAGCATGTACTTCCTCACTTACTTCACGATACTCATCATCATCATGATCATCACTAGCGTCGGCGTCCTGATTCTGGTGAGTCATGGGAATGCTGAGGAAATCAAGGATTTAGTGATTTGA
Protein
MIRDPYKLYVMIFLPIISCALGLYMKSRQIVFFRMQPLTLGPDAYFNKTPIALHSEQNTLREDFKESLEILGAHPIVDFDGNFSSLLDMENFGAFSLKESLNPYGKVMAYYNTTYTHSLPIIINLLDNSLYRALMSASGLEHHFKPIEVMAHPFQQTEQQEEFNLGNVICAVFVGMIFALVPVTLAVDIVYDREIKAKNQLRVNGLSMSMYFLTYFTILIIIMIITSVGVLILVSHGNAEEIKDLVI
Summary
Uniprot
H9JTC7
A0A2H1WF16
A0A3S2LJA8
A0A2A4IY78
A0A2W1BQ17
A0A2H1V1C0
+ More
A0A194Q201 A0A067R5U3 A0A0L7LNT4 A0A2J7PMF3 A0A2J7PME9 D2A4C6 A0A1Y1ME33 A0A2P8ZKQ4 A0A0J7KX87 A0A2A3E528 A0A0T6BE97 A0A088A6H6 N6TSQ4 E9IK73 A0A0M8ZPX7 U4U0E3 F4WYL0 A0A195F057 A0A026WAC0 A0A151JMM8 A0A158P427 A0A195BY13 A0A151XGD7 A0A0L7R8K5 E2BWV8 A0A3L8DZH5 A0A195CTJ9 A0A0U9HY77 B0WRK5 Q17D43 A0A0C9R779 A0A0C9QXZ8 A0A232EPW5 A0A154PKS0 K7IRX5 A0A2S2PMD0 A0A2H8TV16 J9K233 A0A2S2Q7R5 A0A2S2QV61 A0A0A7AS68 A0A1B6D564 A0A1B6CH43 A0A1B6DCT2 A0A1B6DZ36 A0A1B6HSL0 A0A1B6IV27 A0A1B6L4N1 A0A1B6L2P7 A0A0P5Q0E4 A0A0P5NHC6 A0A0P5SAB4 A0A0N7ZT59 A0A0N8EK62 A0A0P5SCK2 A0A0P5FVZ1 A0A0P5M946 A0A0P5NEX5 A0A0P6A7S8 A0A0N8A741 A0A0N8CGI5 A0A0P5QGE7 A0A0P5V6X4 A0A0P6FNG6 A0A0N8AU30 A0A0P6BYZ9 A0A0P6G9I9 A0A0P5DKF7 A0A0P5LAR9
A0A194Q201 A0A067R5U3 A0A0L7LNT4 A0A2J7PMF3 A0A2J7PME9 D2A4C6 A0A1Y1ME33 A0A2P8ZKQ4 A0A0J7KX87 A0A2A3E528 A0A0T6BE97 A0A088A6H6 N6TSQ4 E9IK73 A0A0M8ZPX7 U4U0E3 F4WYL0 A0A195F057 A0A026WAC0 A0A151JMM8 A0A158P427 A0A195BY13 A0A151XGD7 A0A0L7R8K5 E2BWV8 A0A3L8DZH5 A0A195CTJ9 A0A0U9HY77 B0WRK5 Q17D43 A0A0C9R779 A0A0C9QXZ8 A0A232EPW5 A0A154PKS0 K7IRX5 A0A2S2PMD0 A0A2H8TV16 J9K233 A0A2S2Q7R5 A0A2S2QV61 A0A0A7AS68 A0A1B6D564 A0A1B6CH43 A0A1B6DCT2 A0A1B6DZ36 A0A1B6HSL0 A0A1B6IV27 A0A1B6L4N1 A0A1B6L2P7 A0A0P5Q0E4 A0A0P5NHC6 A0A0P5SAB4 A0A0N7ZT59 A0A0N8EK62 A0A0P5SCK2 A0A0P5FVZ1 A0A0P5M946 A0A0P5NEX5 A0A0P6A7S8 A0A0N8A741 A0A0N8CGI5 A0A0P5QGE7 A0A0P5V6X4 A0A0P6FNG6 A0A0N8AU30 A0A0P6BYZ9 A0A0P6G9I9 A0A0P5DKF7 A0A0P5LAR9
Pubmed
EMBL
BABH01004135
BABH01004136
BABH01004137
BABH01004138
ODYU01008243
SOQ51663.1
+ More
RSAL01000093 RVE47906.1 NWSH01005566 PCG64122.1 KZ149935 PZC77159.1 ODYU01000187 SOQ34556.1 KQ459582 KPI99024.1 KK852677 KDR18677.1 JTDY01000471 KOB77024.1 NEVH01024021 PNF17507.1 PNF17508.1 KQ971348 EFA05649.1 GEZM01033856 JAV84079.1 PYGN01000027 PSN57079.1 LBMM01002389 KMQ94889.1 KZ288405 PBC26181.1 LJIG01001320 KRT85620.1 APGK01053503 APGK01053504 KB741224 ENN72300.1 GL763924 EFZ19007.1 KQ435922 KOX68527.1 KB631749 ERL85768.1 GL888453 EGI60698.1 KQ981880 KYN33955.1 KK107323 EZA52591.1 KQ978901 KYN27579.1 ADTU01008287 ADTU01008288 ADTU01008289 ADTU01008290 KQ976394 KYM93185.1 KQ982174 KYQ59358.1 KQ414632 KOC67217.1 GL451188 EFN79822.1 QOIP01000002 RLU25880.1 KQ977304 KYN03817.1 GARF01000030 JAC88913.1 DS232056 EDS33403.1 CH477300 EAT44278.1 GBYB01008657 JAG78424.1 GBYB01008659 JAG78426.1 NNAY01002890 OXU20378.1 KQ434936 KZC11898.1 GGMR01017992 MBY30611.1 GFXV01006300 MBW18105.1 ABLF02039796 ABLF02039804 ABLF02039828 GGMS01004566 MBY73769.1 GGMS01011819 MBY81022.1 KF906265 AHK05630.1 GEDC01016457 JAS20841.1 GEDC01024472 JAS12826.1 GEDC01013819 JAS23479.1 GEDC01006379 JAS30919.1 GECU01030039 JAS77667.1 GECU01016973 JAS90733.1 GEBQ01021298 JAT18679.1 GEBQ01022051 JAT17926.1 GDIQ01121512 JAL30214.1 GDIQ01145258 JAL06468.1 GDIP01142369 JAL61345.1 GDIP01215065 JAJ08337.1 GDIQ01018772 JAN75965.1 GDIQ01106220 JAL45506.1 GDIQ01249576 JAK02149.1 GDIQ01158895 JAK92830.1 GDIQ01146565 JAL05161.1 GDIP01038133 JAM65582.1 GDIP01176009 JAJ47393.1 GDIP01133967 JAL69747.1 GDIQ01135660 JAL16066.1 GDIP01103969 JAL99745.1 GDIQ01056735 JAN38002.1 GDIQ01242819 JAK08906.1 GDIP01012164 JAM91551.1 GDIQ01036651 JAN58086.1 GDIP01171909 JAJ51493.1 GDIQ01172590 JAK79135.1
RSAL01000093 RVE47906.1 NWSH01005566 PCG64122.1 KZ149935 PZC77159.1 ODYU01000187 SOQ34556.1 KQ459582 KPI99024.1 KK852677 KDR18677.1 JTDY01000471 KOB77024.1 NEVH01024021 PNF17507.1 PNF17508.1 KQ971348 EFA05649.1 GEZM01033856 JAV84079.1 PYGN01000027 PSN57079.1 LBMM01002389 KMQ94889.1 KZ288405 PBC26181.1 LJIG01001320 KRT85620.1 APGK01053503 APGK01053504 KB741224 ENN72300.1 GL763924 EFZ19007.1 KQ435922 KOX68527.1 KB631749 ERL85768.1 GL888453 EGI60698.1 KQ981880 KYN33955.1 KK107323 EZA52591.1 KQ978901 KYN27579.1 ADTU01008287 ADTU01008288 ADTU01008289 ADTU01008290 KQ976394 KYM93185.1 KQ982174 KYQ59358.1 KQ414632 KOC67217.1 GL451188 EFN79822.1 QOIP01000002 RLU25880.1 KQ977304 KYN03817.1 GARF01000030 JAC88913.1 DS232056 EDS33403.1 CH477300 EAT44278.1 GBYB01008657 JAG78424.1 GBYB01008659 JAG78426.1 NNAY01002890 OXU20378.1 KQ434936 KZC11898.1 GGMR01017992 MBY30611.1 GFXV01006300 MBW18105.1 ABLF02039796 ABLF02039804 ABLF02039828 GGMS01004566 MBY73769.1 GGMS01011819 MBY81022.1 KF906265 AHK05630.1 GEDC01016457 JAS20841.1 GEDC01024472 JAS12826.1 GEDC01013819 JAS23479.1 GEDC01006379 JAS30919.1 GECU01030039 JAS77667.1 GECU01016973 JAS90733.1 GEBQ01021298 JAT18679.1 GEBQ01022051 JAT17926.1 GDIQ01121512 JAL30214.1 GDIQ01145258 JAL06468.1 GDIP01142369 JAL61345.1 GDIP01215065 JAJ08337.1 GDIQ01018772 JAN75965.1 GDIQ01106220 JAL45506.1 GDIQ01249576 JAK02149.1 GDIQ01158895 JAK92830.1 GDIQ01146565 JAL05161.1 GDIP01038133 JAM65582.1 GDIP01176009 JAJ47393.1 GDIP01133967 JAL69747.1 GDIQ01135660 JAL16066.1 GDIP01103969 JAL99745.1 GDIQ01056735 JAN38002.1 GDIQ01242819 JAK08906.1 GDIP01012164 JAM91551.1 GDIQ01036651 JAN58086.1 GDIP01171909 JAJ51493.1 GDIQ01172590 JAK79135.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000027135
UP000037510
+ More
UP000235965 UP000007266 UP000245037 UP000036403 UP000242457 UP000005203 UP000019118 UP000053105 UP000030742 UP000007755 UP000078541 UP000053097 UP000078492 UP000005205 UP000078540 UP000075809 UP000053825 UP000008237 UP000279307 UP000078542 UP000002320 UP000008820 UP000215335 UP000076502 UP000002358 UP000007819
UP000235965 UP000007266 UP000245037 UP000036403 UP000242457 UP000005203 UP000019118 UP000053105 UP000030742 UP000007755 UP000078541 UP000053097 UP000078492 UP000005205 UP000078540 UP000075809 UP000053825 UP000008237 UP000279307 UP000078542 UP000002320 UP000008820 UP000215335 UP000076502 UP000002358 UP000007819
PRIDE
Pfam
PF00005 ABC_tran
Interpro
CDD
ProteinModelPortal
H9JTC7
A0A2H1WF16
A0A3S2LJA8
A0A2A4IY78
A0A2W1BQ17
A0A2H1V1C0
+ More
A0A194Q201 A0A067R5U3 A0A0L7LNT4 A0A2J7PMF3 A0A2J7PME9 D2A4C6 A0A1Y1ME33 A0A2P8ZKQ4 A0A0J7KX87 A0A2A3E528 A0A0T6BE97 A0A088A6H6 N6TSQ4 E9IK73 A0A0M8ZPX7 U4U0E3 F4WYL0 A0A195F057 A0A026WAC0 A0A151JMM8 A0A158P427 A0A195BY13 A0A151XGD7 A0A0L7R8K5 E2BWV8 A0A3L8DZH5 A0A195CTJ9 A0A0U9HY77 B0WRK5 Q17D43 A0A0C9R779 A0A0C9QXZ8 A0A232EPW5 A0A154PKS0 K7IRX5 A0A2S2PMD0 A0A2H8TV16 J9K233 A0A2S2Q7R5 A0A2S2QV61 A0A0A7AS68 A0A1B6D564 A0A1B6CH43 A0A1B6DCT2 A0A1B6DZ36 A0A1B6HSL0 A0A1B6IV27 A0A1B6L4N1 A0A1B6L2P7 A0A0P5Q0E4 A0A0P5NHC6 A0A0P5SAB4 A0A0N7ZT59 A0A0N8EK62 A0A0P5SCK2 A0A0P5FVZ1 A0A0P5M946 A0A0P5NEX5 A0A0P6A7S8 A0A0N8A741 A0A0N8CGI5 A0A0P5QGE7 A0A0P5V6X4 A0A0P6FNG6 A0A0N8AU30 A0A0P6BYZ9 A0A0P6G9I9 A0A0P5DKF7 A0A0P5LAR9
A0A194Q201 A0A067R5U3 A0A0L7LNT4 A0A2J7PMF3 A0A2J7PME9 D2A4C6 A0A1Y1ME33 A0A2P8ZKQ4 A0A0J7KX87 A0A2A3E528 A0A0T6BE97 A0A088A6H6 N6TSQ4 E9IK73 A0A0M8ZPX7 U4U0E3 F4WYL0 A0A195F057 A0A026WAC0 A0A151JMM8 A0A158P427 A0A195BY13 A0A151XGD7 A0A0L7R8K5 E2BWV8 A0A3L8DZH5 A0A195CTJ9 A0A0U9HY77 B0WRK5 Q17D43 A0A0C9R779 A0A0C9QXZ8 A0A232EPW5 A0A154PKS0 K7IRX5 A0A2S2PMD0 A0A2H8TV16 J9K233 A0A2S2Q7R5 A0A2S2QV61 A0A0A7AS68 A0A1B6D564 A0A1B6CH43 A0A1B6DCT2 A0A1B6DZ36 A0A1B6HSL0 A0A1B6IV27 A0A1B6L4N1 A0A1B6L2P7 A0A0P5Q0E4 A0A0P5NHC6 A0A0P5SAB4 A0A0N7ZT59 A0A0N8EK62 A0A0P5SCK2 A0A0P5FVZ1 A0A0P5M946 A0A0P5NEX5 A0A0P6A7S8 A0A0N8A741 A0A0N8CGI5 A0A0P5QGE7 A0A0P5V6X4 A0A0P6FNG6 A0A0N8AU30 A0A0P6BYZ9 A0A0P6G9I9 A0A0P5DKF7 A0A0P5LAR9
PDB
5XJY
E-value=0.00905856,
Score=89
Ontologies
PANTHER
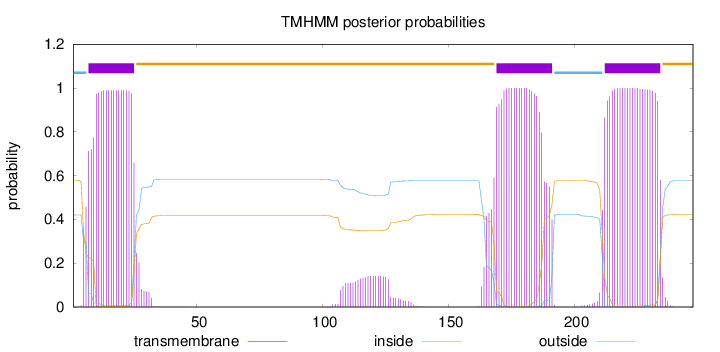
Topology
Length:
247
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
67.81371
Exp number, first 60 AAs:
19.2783
Total prob of N-in:
0.42061
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 25
outside
26 - 168
TMhelix
169 - 191
inside
192 - 211
TMhelix
212 - 234
outside
235 - 247
Population Genetic Test Statistics
Pi
187.025426
Theta
145.370813
Tajima's D
0.965433
CLR
0.122057
CSRT
0.651417429128544
Interpretation
Uncertain
