Gene
KWMTBOMO09456
Pre Gene Modal
BGIBMGA012788
Annotation
Collagenase_[Papilio_machaon]
Full name
Acrosin
Location in the cell
Extracellular Reliability : 2.093
Sequence
CDS
ATGGCACAGTGGGCGGTTTTGCTTCTGGTAGCGATTGCTGGAGTGGCTGTGGTGTCTGCGGATCCGGCTCTGACCTTCGTCGAAAATGTCAGGGCCGGCGCCAGGATCGTATCAGGATGGGAAGCAGAAGAAGGACAGTTCCCTTACCAGCTGTCACTGCGCATGGTGAACCCTGAGGGAGCAGTCAACGCTTGTGGAGCCACCATCATCCACAGCGATTGGGGACTCACGGCTGCTCATTGCACTGCTACGTAA
Protein
MAQWAVLLLVAIAGVAVVSADPALTFVENVRAGARIVSGWEAEEGQFPYQLSLRMVNPEGAVNACGATIIHSDWGLTAAHCTAT
Summary
Description
Acrosin is the major protease of mammalian spermatozoa. It is a serine protease of trypsin-like cleavage specificity, it is synthesized in a zymogen form, proacrosin and stored in the acrosome.
Catalytic Activity
Preferential cleavage: Arg-|-Xaa, Lys-|-Xaa.
Subunit
Heavy chain (catalytic) and a light chain linked by two disulfide bonds.
Similarity
Belongs to the peptidase S1 family.
Feature
chain Acrosin
Uniprot
A0A2H1VYC8
A0A2H1VXG7
A0A0N0PCI5
A0A194Q123
E7D010
J7FDV5
+ More
A0A2A4K409 A0A2A4K3D4 A0A2A4K426 A0A2A4K3B1 B1NLE3 A0A2W1BQ29 A0A2A4K2C1 H9JTC5 A0A2W1BZC6 Q9XY10 B6D1R0 A0A2A4K2C9 J7FCI3 A0A2A4K3S1 B6D1Q9 A0A410KK64 C9W8I8 A0A2H1V7W1 A0A3S2LZV4 A0A212FMW9 A0A2W1BS57 A0A2H1V7Z9 A0A0X9I156 C9W8F4 A0A212F859 A0A2A4K4P0 T1QDG4 A0A194Q024 J7FDE1 A0A194RS93 A0PGD9 A0A0K8S2W5 A0A2W1BWI0 A0A3S2NJF1 B4X992 A0A2H1VV70 A0A437BBW9 H9JTB5 A0A2H1VV25 A0A2A4K3Y4 A0A109P6B5 A0A2H1WBT2 Q296D1 B4GFB5 A0A3Q2XDK3 T1GQY2 B6D5J3
A0A2A4K409 A0A2A4K3D4 A0A2A4K426 A0A2A4K3B1 B1NLE3 A0A2W1BQ29 A0A2A4K2C1 H9JTC5 A0A2W1BZC6 Q9XY10 B6D1R0 A0A2A4K2C9 J7FCI3 A0A2A4K3S1 B6D1Q9 A0A410KK64 C9W8I8 A0A2H1V7W1 A0A3S2LZV4 A0A212FMW9 A0A2W1BS57 A0A2H1V7Z9 A0A0X9I156 C9W8F4 A0A212F859 A0A2A4K4P0 T1QDG4 A0A194Q024 J7FDE1 A0A194RS93 A0PGD9 A0A0K8S2W5 A0A2W1BWI0 A0A3S2NJF1 B4X992 A0A2H1VV70 A0A437BBW9 H9JTB5 A0A2H1VV25 A0A2A4K3Y4 A0A109P6B5 A0A2H1WBT2 Q296D1 B4GFB5 A0A3Q2XDK3 T1GQY2 B6D5J3
EC Number
3.4.21.10
Pubmed
EMBL
ODYU01005031
SOQ45512.1
ODYU01005032
SOQ45513.1
KQ460651
KPJ13055.1
+ More
KQ459582 KPI99023.1 HM990180 ADT80828.1 JN252040 AFM28253.1 NWSH01000207 PCG78410.1 PCG78414.1 PCG78412.1 PCG78408.1 EF600058 ABU98623.1 KZ149935 PZC77162.1 PCG78411.1 BABH01004141 PZC77163.1 AB026735 BAA77401.1 EU673453 ACI45416.1 PCG78415.1 JN252039 AFM28252.1 PCG78413.1 EU673452 ACI45415.1 MH201547 QAR48640.1 FJ205435 ACR16001.2 ODYU01001144 SOQ36940.1 RSAL01000093 RVE47905.1 AGBW02007651 OWR55096.1 PZC77161.1 ODYU01001143 SOQ36937.1 KP690084 ALO61086.1 FJ205398 ACR15967.2 AGBW02009787 OWR49920.1 NWSH01000176 PCG78733.1 JQ429439 AFW03964.1 KPI98932.1 JN252043 AFM28256.1 KQ459700 KPJ20347.1 AY618894 AAV33657.1 GBRD01018206 JAG47621.1 PZC77160.1 RSAL01000071 RVE49104.1 EF531628 ABR88239.1 ODYU01004626 SOQ44698.1 RVE47909.1 BABH01004171 BABH01004172 SOQ44697.1 PCG78736.1 KP690083 ALO61085.1 ODYU01007484 SOQ50292.1 CM000070 EAL28527.2 CH479182 EDW34300.1 CAQQ02394015 EU836256 ACI22824.1
KQ459582 KPI99023.1 HM990180 ADT80828.1 JN252040 AFM28253.1 NWSH01000207 PCG78410.1 PCG78414.1 PCG78412.1 PCG78408.1 EF600058 ABU98623.1 KZ149935 PZC77162.1 PCG78411.1 BABH01004141 PZC77163.1 AB026735 BAA77401.1 EU673453 ACI45416.1 PCG78415.1 JN252039 AFM28252.1 PCG78413.1 EU673452 ACI45415.1 MH201547 QAR48640.1 FJ205435 ACR16001.2 ODYU01001144 SOQ36940.1 RSAL01000093 RVE47905.1 AGBW02007651 OWR55096.1 PZC77161.1 ODYU01001143 SOQ36937.1 KP690084 ALO61086.1 FJ205398 ACR15967.2 AGBW02009787 OWR49920.1 NWSH01000176 PCG78733.1 JQ429439 AFW03964.1 KPI98932.1 JN252043 AFM28256.1 KQ459700 KPJ20347.1 AY618894 AAV33657.1 GBRD01018206 JAG47621.1 PZC77160.1 RSAL01000071 RVE49104.1 EF531628 ABR88239.1 ODYU01004626 SOQ44698.1 RVE47909.1 BABH01004171 BABH01004172 SOQ44697.1 PCG78736.1 KP690083 ALO61085.1 ODYU01007484 SOQ50292.1 CM000070 EAL28527.2 CH479182 EDW34300.1 CAQQ02394015 EU836256 ACI22824.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
A0A2H1VYC8
A0A2H1VXG7
A0A0N0PCI5
A0A194Q123
E7D010
J7FDV5
+ More
A0A2A4K409 A0A2A4K3D4 A0A2A4K426 A0A2A4K3B1 B1NLE3 A0A2W1BQ29 A0A2A4K2C1 H9JTC5 A0A2W1BZC6 Q9XY10 B6D1R0 A0A2A4K2C9 J7FCI3 A0A2A4K3S1 B6D1Q9 A0A410KK64 C9W8I8 A0A2H1V7W1 A0A3S2LZV4 A0A212FMW9 A0A2W1BS57 A0A2H1V7Z9 A0A0X9I156 C9W8F4 A0A212F859 A0A2A4K4P0 T1QDG4 A0A194Q024 J7FDE1 A0A194RS93 A0PGD9 A0A0K8S2W5 A0A2W1BWI0 A0A3S2NJF1 B4X992 A0A2H1VV70 A0A437BBW9 H9JTB5 A0A2H1VV25 A0A2A4K3Y4 A0A109P6B5 A0A2H1WBT2 Q296D1 B4GFB5 A0A3Q2XDK3 T1GQY2 B6D5J3
A0A2A4K409 A0A2A4K3D4 A0A2A4K426 A0A2A4K3B1 B1NLE3 A0A2W1BQ29 A0A2A4K2C1 H9JTC5 A0A2W1BZC6 Q9XY10 B6D1R0 A0A2A4K2C9 J7FCI3 A0A2A4K3S1 B6D1Q9 A0A410KK64 C9W8I8 A0A2H1V7W1 A0A3S2LZV4 A0A212FMW9 A0A2W1BS57 A0A2H1V7Z9 A0A0X9I156 C9W8F4 A0A212F859 A0A2A4K4P0 T1QDG4 A0A194Q024 J7FDE1 A0A194RS93 A0PGD9 A0A0K8S2W5 A0A2W1BWI0 A0A3S2NJF1 B4X992 A0A2H1VV70 A0A437BBW9 H9JTB5 A0A2H1VV25 A0A2A4K3Y4 A0A109P6B5 A0A2H1WBT2 Q296D1 B4GFB5 A0A3Q2XDK3 T1GQY2 B6D5J3
PDB
3HKI
E-value=1.01506e-05,
Score=110
Ontologies
GO
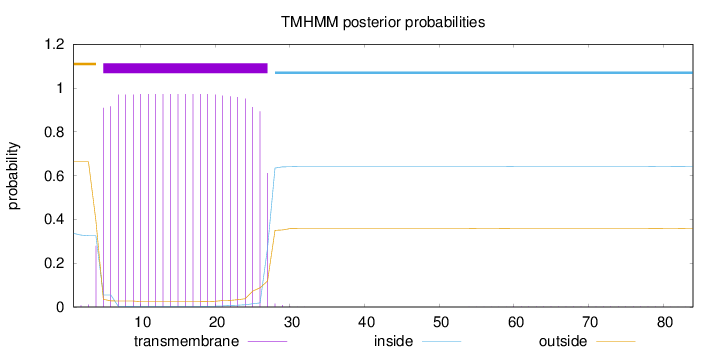
Topology
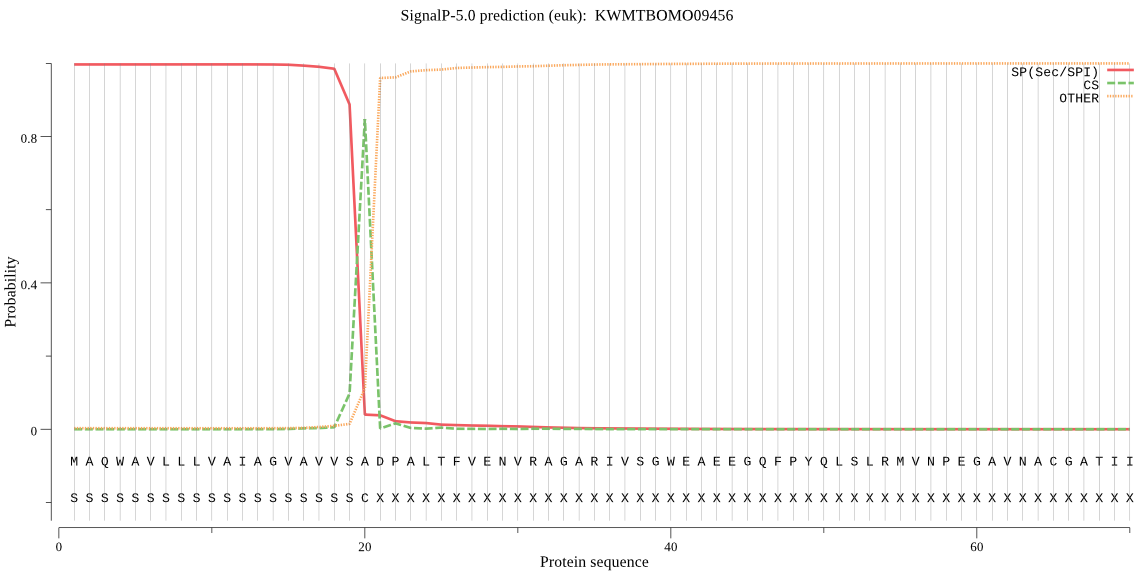
SignalP
Position: 1 - 20,
Likelihood: 0.996821
Length:
84
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.06282
Exp number, first 60 AAs:
22.01811
Total prob of N-in:
0.33626
POSSIBLE N-term signal
sequence
outside
1 - 4
TMhelix
5 - 27
inside
28 - 84
Population Genetic Test Statistics
Pi
188.979766
Theta
147.35863
Tajima's D
0.816871
CLR
2.874074
CSRT
0.601069946502675
Interpretation
Uncertain
