Gene
KWMTBOMO09454
Pre Gene Modal
BGIBMGA012788
Annotation
PREDICTED:_mucin-2-like_isoform_X1_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.038 Nuclear Reliability : 1.019
Sequence
CDS
ATGGCCGGGCGATTGAGTGGTCGTTATGCGTTCGATATTAAAAAGTGGGCGCTCGCCGACGACACCGATTTCACTTTCCCTGAAATCGCTCGCGAGCGTTCCCTGCCGGGTTCTAGAATTGTATCAGGATGGGAGGCGAGTGAAGGCCAGTTCCCTTACCAGCTCTCGATTCGTATGGTCAGTACTGTGGGAGGAGTCAACGCCTGCGGAGCCACCATCATTCATAGCAACTGGGGGCTTACAGCTGCTCACTGCACCGGTTTGCGTGTAACAATCATCGTCCGAGCTGGAGCAGTGAACCTGACCCGTCCCGGCTTGCTTTTCGAGACCACTAAGTACATAAATCATCCTGAGTACAGCGAAAATTTGAACGTAGTGCAGCCTCATGATATTGGTCTAATAGACTTTGGTAGAAAACTAGAGTTTAATGATTACATTCAGCCAATCAGACTGCAGAGATCCGCGGATAAAGATCGTAATTACGACAATGTCAGACTTGTCGCCAGCGGATGGGGCCGCACCTGGACCGGAAGCGCATCTCCGGAGAACCTGAACTGGGTGTTCTTGAACGGAATATCGAATCTGCGTTGTATGGTGGCCTACAACTTCAGTCCCACCATTCAGCCCTCCACCATTTGCACGCTTGGTTACAACGACACCACGCAGTCCACTTGCCAGGGTGACAGCGGTGGTCCTCTGACAGTGATCGATGAAGACGGCCAAATCACACAAGTAGGAGTCACTTCGTTCGTGTCCAGCGAAGGCTGCCACGTGGATATACCTGCAGGTTTTATCCGGCCCGGCCACTACCTCGACTGGTTTAAGACGGTCACCGGTTTAGATTTCGATTGGACATCGAGCACAACACAAGCACCTGAGACAACAGAAACACCCGGCGCAACAGAAGCGCCTGTGACAACAGAAGCTCCCGAGGCCACAGAAGCACCCGTGACCACAGAAGCACCCGGCGCAACAGAAGCACCCAACACAACTGAAGCACCCGACGCAACAGAAGCACCCGTGACCACAGAAGCACCCGAGATAACAGAAGCACCTTACACAACAGAAACACCCGAGACCCCAGAAACCAGATACCGAAACTTCGGCACCGCTTTTTGA
Protein
MAGRLSGRYAFDIKKWALADDTDFTFPEIARERSLPGSRIVSGWEASEGQFPYQLSIRMVSTVGGVNACGATIIHSNWGLTAAHCTGLRVTIIVRAGAVNLTRPGLLFETTKYINHPEYSENLNVVQPHDIGLIDFGRKLEFNDYIQPIRLQRSADKDRNYDNVRLVASGWGRTWTGSASPENLNWVFLNGISNLRCMVAYNFSPTIQPSTICTLGYNDTTQSTCQGDSGGPLTVIDEDGQITQVGVTSFVSSEGCHVDIPAGFIRPGHYLDWFKTVTGLDFDWTSSTTQAPETTETPGATEAPVTTEAPEATEAPVTTEAPGATEAPNTTEAPDATEAPVTTEAPEITEAPYTTETPETPETRYRNFGTAF
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
Q9XY10
H9JTC6
E7D010
A0A3S2LZV4
A0A2A4K409
A0A2A4K3D4
+ More
A0A2A4K426 J7FDV5 A0A410KK64 A0A2A4K2C1 A0A2A4K3S1 A0A2A4K2C9 B1NLE3 A0A2W1BQ29 C9W8I8 A0A2A4K3B1 B6D1R0 B6D1Q9 A0A2W1BZC6 A0A194Q123 J7FDE1 B4X992 H9JTD0 C9W8H1 A0A0X9I156 A0A2H1VXD9 A0A2H1V7Z9 A0A2A4K3Y4 C9W8F4 A0A212F859 A0A2W1BS57 A0A2A4K4P0 A0A0N1IP15 A0A2H1WBT2 A0A109P6B5 C9W8G6 A0A2H1VV25 H9JTC9 Q52NW5 A0A182SLA6 A0A182RAB0 A0A182MAA4 A0A084W5I8 A0A182IN32 A0A182PK66 A0A1I8P095 B0WD43
A0A2A4K426 J7FDV5 A0A410KK64 A0A2A4K2C1 A0A2A4K3S1 A0A2A4K2C9 B1NLE3 A0A2W1BQ29 C9W8I8 A0A2A4K3B1 B6D1R0 B6D1Q9 A0A2W1BZC6 A0A194Q123 J7FDE1 B4X992 H9JTD0 C9W8H1 A0A0X9I156 A0A2H1VXD9 A0A2H1V7Z9 A0A2A4K3Y4 C9W8F4 A0A212F859 A0A2W1BS57 A0A2A4K4P0 A0A0N1IP15 A0A2H1WBT2 A0A109P6B5 C9W8G6 A0A2H1VV25 H9JTC9 Q52NW5 A0A182SLA6 A0A182RAB0 A0A182MAA4 A0A084W5I8 A0A182IN32 A0A182PK66 A0A1I8P095 B0WD43
Pubmed
EMBL
AB026735
BAA77401.1
BABH01004139
BABH01004140
BABH01004141
HM990180
+ More
ADT80828.1 RSAL01000093 RVE47905.1 NWSH01000207 PCG78410.1 PCG78414.1 PCG78412.1 JN252040 AFM28253.1 MH201547 QAR48640.1 PCG78411.1 PCG78413.1 PCG78415.1 EF600058 ABU98623.1 KZ149935 PZC77162.1 FJ205435 ACR16001.2 PCG78408.1 EU673453 ACI45416.1 EU673452 ACI45415.1 PZC77163.1 KQ459582 KPI99023.1 JN252043 AFM28256.1 EF531628 ABR88239.1 BABH01004119 BABH01004120 BABH01004121 BABH01004122 FJ205416 ACR15984.1 KP690084 ALO61086.1 ODYU01005031 SOQ45511.1 ODYU01001143 SOQ36937.1 NWSH01000176 PCG78736.1 FJ205398 ACR15967.2 AGBW02009787 OWR49920.1 PZC77161.1 PCG78733.1 KQ460651 KPJ13048.1 ODYU01007484 SOQ50292.1 KP690083 ALO61085.1 FJ205411 ACR15979.2 ODYU01004626 SOQ44697.1 BABH01004123 BABH01004124 AY995188 AAX94742.1 AXCM01001005 ATLV01020656 KE525304 KFB45482.1 DS231893 EDS44178.1
ADT80828.1 RSAL01000093 RVE47905.1 NWSH01000207 PCG78410.1 PCG78414.1 PCG78412.1 JN252040 AFM28253.1 MH201547 QAR48640.1 PCG78411.1 PCG78413.1 PCG78415.1 EF600058 ABU98623.1 KZ149935 PZC77162.1 FJ205435 ACR16001.2 PCG78408.1 EU673453 ACI45416.1 EU673452 ACI45415.1 PZC77163.1 KQ459582 KPI99023.1 JN252043 AFM28256.1 EF531628 ABR88239.1 BABH01004119 BABH01004120 BABH01004121 BABH01004122 FJ205416 ACR15984.1 KP690084 ALO61086.1 ODYU01005031 SOQ45511.1 ODYU01001143 SOQ36937.1 NWSH01000176 PCG78736.1 FJ205398 ACR15967.2 AGBW02009787 OWR49920.1 PZC77161.1 PCG78733.1 KQ460651 KPJ13048.1 ODYU01007484 SOQ50292.1 KP690083 ALO61085.1 FJ205411 ACR15979.2 ODYU01004626 SOQ44697.1 BABH01004123 BABH01004124 AY995188 AAX94742.1 AXCM01001005 ATLV01020656 KE525304 KFB45482.1 DS231893 EDS44178.1
Proteomes
Pfam
PF00089 Trypsin
Interpro
SUPFAM
SSF50494
SSF50494
CDD
ProteinModelPortal
Q9XY10
H9JTC6
E7D010
A0A3S2LZV4
A0A2A4K409
A0A2A4K3D4
+ More
A0A2A4K426 J7FDV5 A0A410KK64 A0A2A4K2C1 A0A2A4K3S1 A0A2A4K2C9 B1NLE3 A0A2W1BQ29 C9W8I8 A0A2A4K3B1 B6D1R0 B6D1Q9 A0A2W1BZC6 A0A194Q123 J7FDE1 B4X992 H9JTD0 C9W8H1 A0A0X9I156 A0A2H1VXD9 A0A2H1V7Z9 A0A2A4K3Y4 C9W8F4 A0A212F859 A0A2W1BS57 A0A2A4K4P0 A0A0N1IP15 A0A2H1WBT2 A0A109P6B5 C9W8G6 A0A2H1VV25 H9JTC9 Q52NW5 A0A182SLA6 A0A182RAB0 A0A182MAA4 A0A084W5I8 A0A182IN32 A0A182PK66 A0A1I8P095 B0WD43
A0A2A4K426 J7FDV5 A0A410KK64 A0A2A4K2C1 A0A2A4K3S1 A0A2A4K2C9 B1NLE3 A0A2W1BQ29 C9W8I8 A0A2A4K3B1 B6D1R0 B6D1Q9 A0A2W1BZC6 A0A194Q123 J7FDE1 B4X992 H9JTD0 C9W8H1 A0A0X9I156 A0A2H1VXD9 A0A2H1V7Z9 A0A2A4K3Y4 C9W8F4 A0A212F859 A0A2W1BS57 A0A2A4K4P0 A0A0N1IP15 A0A2H1WBT2 A0A109P6B5 C9W8G6 A0A2H1VV25 H9JTC9 Q52NW5 A0A182SLA6 A0A182RAB0 A0A182MAA4 A0A084W5I8 A0A182IN32 A0A182PK66 A0A1I8P095 B0WD43
PDB
2HLC
E-value=3.06379e-27,
Score=303
Ontologies
GO
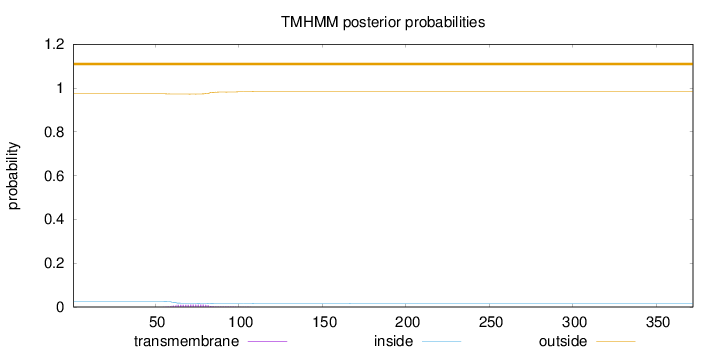
Topology
Length:
372
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.26949
Exp number, first 60 AAs:
0.01282
Total prob of N-in:
0.02643
outside
1 - 372
Population Genetic Test Statistics
Pi
200.803265
Theta
152.231681
Tajima's D
0.118669
CLR
0.581239
CSRT
0.405679716014199
Interpretation
Uncertain
