Gene
KWMTBOMO09451 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013072
Annotation
PREDICTED:_leucine-rich_repeat-containing_protein_47-like_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 3.85
Sequence
CDS
ATGAGTGCTTGGCCGGAAGTTGTGACCGCAAAAGAGGAGAATCGTCACGAAATTAAGTTGGCGGGAGCCGCTATATCGAAACGAATCTCTGAACATGGCTTAGACAAAAATATCTTCACCTTAACAAACATAAACTTATTAAACATCAGTGATACATGTCTGCACGCTATACCAGATGAAATCAAGAACTTAATTAATCTCCAAAGCTTGTTAATGTTTGGAAATAAGATTGAAGTTTTTAATGAAAACATAACCACACTTCGAAAGTTAAAAGTCCTCGATTTATCAAGAAACGAATTAAAAAGCATCCCAGAGAGCTTGAACAACATGAAAGAACTAACGAGTATCAATTTCAGCTCAAATCAAATAGAAGATATGCCGAAAATTGGCGATTTACCAAACTTAATAATAATAGACATTTCCAATAATAAACTGAAGAGTTTCTTGGACGTGGAATCGGCTAACTTGCCTCATTTGACAGACTTGAAGCTAAAAAGTAATTTGATTGAATCTATTCCAAGTTGTATCGGACAAAAACTTTCATCACTCAAGAACTTTGATATCAGCGACAATCAAATAAATGTTGTACCTGGCGAAGTAGCTAACATGTCGAAAATTAAAGATCTCAACTTAAAGGGGAACAAGCTATCAGACAAGCGGTTCATGAAACTGGTTGATCAGTGTCGCACTAAACAAATTATGGACTACATCCGCGATCACTGCCCCAAAACAGAACAAACTCAACAGTCATCCAAATCCAAAACAAAAAAAGGAAAGAAAGAAGAACCTCATACAGAAAATGTGGAAGCTGAATTGTGTAACTCAATGAAGATTTTACATGTCTCTGACGATGTATTGAACGTTAAAATTGTTGAGTCTCAGGTTGGCAGTGTCCGACCTTATATACTTTGTTGCATAATAAAGGATCTAAATTTCACAGATATATTGTTTAAAAAGTTTATACAAATGCAAACAAGACTGCATGACACCGTCTGTGATAAGAGAAATGTAGCCACAATTGCAACACATGACCTTAACAAATTACCTCCAGGTGATCTTGTGTACACAGCAAAGCCTCCGTCAATGCTAAGCCTTATTCCGTTAATGAGGACAAAACCCTACACCGGAGAACAACTAGTCCAGCAGTTAGTAGCAGAAGCGGAAGCACTCAGGAAGGAGAAAAAAAGAAATGTTTACTCGGGGATCCATAAGTACCTGTACCTGTTAGAAGGCAAAGCAGTCTATGCTTGCCTCCAAGACTCATCCGGTAGAGTAATCAGCTTCCCACCTATCACCAATGCTGACGTAACGAAGATGTCATCAGAAAGTAAAGCCATGCTGATAGAGGTGACATCTCATTCTTCACTCGGAGCCTGTAAGATGGTGATGGATAAAATATTGCAAGAATGTTTGATGCTAGGCATCGGAGCTGACTCCCAGGACGGTTTTCATGCGCTCACTGTGCAACAGGTCAAAGTAGTCGACCCGGATGGTAACCTGAAGTCAGTGTATCCATCGCGAACTGACTGTGTGTATGATGATACGAACATCAAAGTCATCAGGATGCCTAAAAAATAA
Protein
MSAWPEVVTAKEENRHEIKLAGAAISKRISEHGLDKNIFTLTNINLLNISDTCLHAIPDEIKNLINLQSLLMFGNKIEVFNENITTLRKLKVLDLSRNELKSIPESLNNMKELTSINFSSNQIEDMPKIGDLPNLIIIDISNNKLKSFLDVESANLPHLTDLKLKSNLIESIPSCIGQKLSSLKNFDISDNQINVVPGEVANMSKIKDLNLKGNKLSDKRFMKLVDQCRTKQIMDYIRDHCPKTEQTQQSSKSKTKKGKKEEPHTENVEAELCNSMKILHVSDDVLNVKIVESQVGSVRPYILCCIIKDLNFTDILFKKFIQMQTRLHDTVCDKRNVATIATHDLNKLPPGDLVYTAKPPSMLSLIPLMRTKPYTGEQLVQQLVAEAEALRKEKKRNVYSGIHKYLYLLEGKAVYACLQDSSGRVISFPPITNADVTKMSSESKAMLIEVTSHSSLGACKMVMDKILQECLMLGIGADSQDGFHALTVQQVKVVDPDGNLKSVYPSRTDCVYDDTNIKVIRMPKK
Summary
Uniprot
H9JU60
A0A2H1VXF0
A0A2A4K312
A0A194Q0B5
A0A212FMX7
A0A437BC24
+ More
A0A0N1PG54 A0A2J7Q743 A0A067R2N7 A0A2P8Y3F0 A0A026VZW6 A0A2A3EJ94 A0A087ZQ06 A0A151JU34 A0A232F8E1 A0A151IHN0 A0A151I5C9 K7ITZ8 A0A158NWS6 F4WQ34 A0A0M9A5L9 A0A154PFM3 E9IVZ5 A0A0L7QUQ7 A0A195EDC8 A0A310SUS1 A0A1W4XC59 A0A151WY70 A0A1Y1M253 A0A411G7L6 A0A0T6B631 A0A1S3H2L3 D2A5G5 T1J910 A0A087TK97 A0A2H4KB79 V5HD17 A0A131XZC1 A0A293LVL2 A0A2R5LM91 E0VK28 A0A224YVM6 B7QHR9 L7M8B3 A0A131YM47 A0A131XHY6 A0A0P4WT28 G3MPG1 A0A1E1XSV8 A0A210Q877 A0A443R7J4 V4AF39 A0A1A8EX67 A0A1A8CG66 A0A1A8DM65 A0A1A8MRD3 A0A1D1V3R2 A0A1A8UPU1 A0A1W0WWQ4 A0A1A8IIY3 A0A1A7YYD1 A0A1A8S5I9 A0A3Q0SBH2 A0A1E1X6C6 A0A2C9JGF0 A0A085NDM6 A0A3Q3A144 A0A3B3YCA9 A0A2U9BYK1 A0A3B3VDK4 A0A2I4C794 A0A3Q3MAN9 T1KEG9 A0A3P8YPV7 R7TQ29 A0A3Q1HIW5 A0A3Q3J341 A0A3Q1GMK9 A0A315WB05 A0A3B4CLM5 A0A0S7L5Q9 A0A3B5L107 A0A3P8SSQ0 A0A3Q2VXV4 A0A3Q1AUG3 A0A3P8Q9K3 A0A3P9D223 A0A3B5QE97 W4ZCP1 A0A3Q1GX48 A0A293LFE6 I3JHM1 A0A3Q3XGY2 A0A3B4G1L7 A0A3P9P2H6 A0A3B4UP26 A0A3B4YN92
A0A0N1PG54 A0A2J7Q743 A0A067R2N7 A0A2P8Y3F0 A0A026VZW6 A0A2A3EJ94 A0A087ZQ06 A0A151JU34 A0A232F8E1 A0A151IHN0 A0A151I5C9 K7ITZ8 A0A158NWS6 F4WQ34 A0A0M9A5L9 A0A154PFM3 E9IVZ5 A0A0L7QUQ7 A0A195EDC8 A0A310SUS1 A0A1W4XC59 A0A151WY70 A0A1Y1M253 A0A411G7L6 A0A0T6B631 A0A1S3H2L3 D2A5G5 T1J910 A0A087TK97 A0A2H4KB79 V5HD17 A0A131XZC1 A0A293LVL2 A0A2R5LM91 E0VK28 A0A224YVM6 B7QHR9 L7M8B3 A0A131YM47 A0A131XHY6 A0A0P4WT28 G3MPG1 A0A1E1XSV8 A0A210Q877 A0A443R7J4 V4AF39 A0A1A8EX67 A0A1A8CG66 A0A1A8DM65 A0A1A8MRD3 A0A1D1V3R2 A0A1A8UPU1 A0A1W0WWQ4 A0A1A8IIY3 A0A1A7YYD1 A0A1A8S5I9 A0A3Q0SBH2 A0A1E1X6C6 A0A2C9JGF0 A0A085NDM6 A0A3Q3A144 A0A3B3YCA9 A0A2U9BYK1 A0A3B3VDK4 A0A2I4C794 A0A3Q3MAN9 T1KEG9 A0A3P8YPV7 R7TQ29 A0A3Q1HIW5 A0A3Q3J341 A0A3Q1GMK9 A0A315WB05 A0A3B4CLM5 A0A0S7L5Q9 A0A3B5L107 A0A3P8SSQ0 A0A3Q2VXV4 A0A3Q1AUG3 A0A3P8Q9K3 A0A3P9D223 A0A3B5QE97 W4ZCP1 A0A3Q1GX48 A0A293LFE6 I3JHM1 A0A3Q3XGY2 A0A3B4G1L7 A0A3P9P2H6 A0A3B4UP26 A0A3B4YN92
Pubmed
19121390
26354079
22118469
24845553
29403074
24508170
+ More
30249741 28648823 20075255 21347285 21719571 21282665 28004739 18362917 19820115 25765539 20566863 28797301 25576852 26830274 28049606 22216098 29209593 28812685 23254933 27649274 28503490 15562597 24929829 25069045 29703783 25186727 23542700
30249741 28648823 20075255 21347285 21719571 21282665 28004739 18362917 19820115 25765539 20566863 28797301 25576852 26830274 28049606 22216098 29209593 28812685 23254933 27649274 28503490 15562597 24929829 25069045 29703783 25186727 23542700
EMBL
BABH01004143
ODYU01005031
SOQ45510.1
NWSH01000207
PCG78406.1
KQ459582
+ More
KPI99022.1 AGBW02007651 OWR55095.1 RSAL01000093 RVE47904.1 KQ460651 KPJ13056.1 NEVH01017447 PNF24401.1 KK853054 KDR12004.1 PYGN01000977 PSN38760.1 KK107522 QOIP01000001 EZA49348.1 RLU27616.1 KZ288227 PBC31797.1 KQ981799 KYN35696.1 NNAY01000667 OXU27096.1 KQ977586 KYN01687.1 KQ976434 KYM87199.1 ADTU01002979 GL888262 EGI63657.1 KQ435750 KOX76349.1 KQ434893 KZC10612.1 GL766449 EFZ15281.1 KQ414731 KOC62387.1 KQ979074 KYN22854.1 KQ760241 OAD61290.1 KQ982651 KYQ52834.1 GEZM01045089 JAV77926.1 MH366084 QBB01893.1 LJIG01009584 KRT82787.1 KQ971345 EFA05368.1 JH431968 KK115611 KFM65536.1 KX550100 ASR74814.1 GANP01011496 JAB72972.1 GEFM01003202 JAP72594.1 GFWV01007078 MAA31808.1 GGLE01006422 MBY10548.1 DS235237 EEB13734.1 GFPF01010521 MAA21667.1 ABJB010721290 ABJB011098710 DS941498 EEC18391.1 GACK01004957 JAA60077.1 GEDV01008214 JAP80343.1 GEFH01002454 JAP66127.1 GDRN01025029 JAI67735.1 JO843762 AEO35379.1 GFAA01001052 JAU02383.1 NEDP02004651 OWF44940.1 NCKU01001793 RWS11240.1 KB201656 ESO95482.1 HAEB01004594 SBQ51121.1 HADZ01014856 SBP78797.1 HAEA01006153 SBQ34633.1 HAEF01018083 HAEG01003687 SBR59242.1 BDGG01000002 GAU93088.1 HADY01000667 HAEJ01009169 SBS49626.1 MTYJ01000037 OQV19627.1 HAED01010835 HAEE01002120 SBQ97074.1 HADW01014092 HADX01013390 SBP35622.1 HAEI01011240 SBS12843.1 GFAC01004592 JAT94596.1 KL363198 KL367513 KFD55740.1 KFD67572.1 KFD67577.1 CP026253 AWP09334.1 CAEY01002034 AMQN01002628 KB309694 ELT93616.1 NHOQ01000099 PWA33003.1 GBYX01184558 JAO84846.1 AAGJ04054098 AAGJ04054099 GFWV01002733 MAA27463.1 AERX01037014
KPI99022.1 AGBW02007651 OWR55095.1 RSAL01000093 RVE47904.1 KQ460651 KPJ13056.1 NEVH01017447 PNF24401.1 KK853054 KDR12004.1 PYGN01000977 PSN38760.1 KK107522 QOIP01000001 EZA49348.1 RLU27616.1 KZ288227 PBC31797.1 KQ981799 KYN35696.1 NNAY01000667 OXU27096.1 KQ977586 KYN01687.1 KQ976434 KYM87199.1 ADTU01002979 GL888262 EGI63657.1 KQ435750 KOX76349.1 KQ434893 KZC10612.1 GL766449 EFZ15281.1 KQ414731 KOC62387.1 KQ979074 KYN22854.1 KQ760241 OAD61290.1 KQ982651 KYQ52834.1 GEZM01045089 JAV77926.1 MH366084 QBB01893.1 LJIG01009584 KRT82787.1 KQ971345 EFA05368.1 JH431968 KK115611 KFM65536.1 KX550100 ASR74814.1 GANP01011496 JAB72972.1 GEFM01003202 JAP72594.1 GFWV01007078 MAA31808.1 GGLE01006422 MBY10548.1 DS235237 EEB13734.1 GFPF01010521 MAA21667.1 ABJB010721290 ABJB011098710 DS941498 EEC18391.1 GACK01004957 JAA60077.1 GEDV01008214 JAP80343.1 GEFH01002454 JAP66127.1 GDRN01025029 JAI67735.1 JO843762 AEO35379.1 GFAA01001052 JAU02383.1 NEDP02004651 OWF44940.1 NCKU01001793 RWS11240.1 KB201656 ESO95482.1 HAEB01004594 SBQ51121.1 HADZ01014856 SBP78797.1 HAEA01006153 SBQ34633.1 HAEF01018083 HAEG01003687 SBR59242.1 BDGG01000002 GAU93088.1 HADY01000667 HAEJ01009169 SBS49626.1 MTYJ01000037 OQV19627.1 HAED01010835 HAEE01002120 SBQ97074.1 HADW01014092 HADX01013390 SBP35622.1 HAEI01011240 SBS12843.1 GFAC01004592 JAT94596.1 KL363198 KL367513 KFD55740.1 KFD67572.1 KFD67577.1 CP026253 AWP09334.1 CAEY01002034 AMQN01002628 KB309694 ELT93616.1 NHOQ01000099 PWA33003.1 GBYX01184558 JAO84846.1 AAGJ04054098 AAGJ04054099 GFWV01002733 MAA27463.1 AERX01037014
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000283053
UP000053240
+ More
UP000235965 UP000027135 UP000245037 UP000053097 UP000279307 UP000242457 UP000005203 UP000078541 UP000215335 UP000078542 UP000078540 UP000002358 UP000005205 UP000007755 UP000053105 UP000076502 UP000053825 UP000078492 UP000192223 UP000075809 UP000085678 UP000007266 UP000054359 UP000009046 UP000001555 UP000242188 UP000285301 UP000030746 UP000186922 UP000261340 UP000076420 UP000264800 UP000261480 UP000246464 UP000261500 UP000192220 UP000261640 UP000015104 UP000265140 UP000014760 UP000265040 UP000261600 UP000257200 UP000261440 UP000261380 UP000265080 UP000264840 UP000257160 UP000265100 UP000265160 UP000002852 UP000007110 UP000005207 UP000261620 UP000261460 UP000242638 UP000261420 UP000261360
UP000235965 UP000027135 UP000245037 UP000053097 UP000279307 UP000242457 UP000005203 UP000078541 UP000215335 UP000078542 UP000078540 UP000002358 UP000005205 UP000007755 UP000053105 UP000076502 UP000053825 UP000078492 UP000192223 UP000075809 UP000085678 UP000007266 UP000054359 UP000009046 UP000001555 UP000242188 UP000285301 UP000030746 UP000186922 UP000261340 UP000076420 UP000264800 UP000261480 UP000246464 UP000261500 UP000192220 UP000261640 UP000015104 UP000265140 UP000014760 UP000265040 UP000261600 UP000257200 UP000261440 UP000261380 UP000265080 UP000264840 UP000257160 UP000265100 UP000265160 UP000002852 UP000007110 UP000005207 UP000261620 UP000261460 UP000242638 UP000261420 UP000261360
PRIDE
Pfam
Interpro
SUPFAM
SSF52540
SSF52540
Gene 3D
CDD
ProteinModelPortal
H9JU60
A0A2H1VXF0
A0A2A4K312
A0A194Q0B5
A0A212FMX7
A0A437BC24
+ More
A0A0N1PG54 A0A2J7Q743 A0A067R2N7 A0A2P8Y3F0 A0A026VZW6 A0A2A3EJ94 A0A087ZQ06 A0A151JU34 A0A232F8E1 A0A151IHN0 A0A151I5C9 K7ITZ8 A0A158NWS6 F4WQ34 A0A0M9A5L9 A0A154PFM3 E9IVZ5 A0A0L7QUQ7 A0A195EDC8 A0A310SUS1 A0A1W4XC59 A0A151WY70 A0A1Y1M253 A0A411G7L6 A0A0T6B631 A0A1S3H2L3 D2A5G5 T1J910 A0A087TK97 A0A2H4KB79 V5HD17 A0A131XZC1 A0A293LVL2 A0A2R5LM91 E0VK28 A0A224YVM6 B7QHR9 L7M8B3 A0A131YM47 A0A131XHY6 A0A0P4WT28 G3MPG1 A0A1E1XSV8 A0A210Q877 A0A443R7J4 V4AF39 A0A1A8EX67 A0A1A8CG66 A0A1A8DM65 A0A1A8MRD3 A0A1D1V3R2 A0A1A8UPU1 A0A1W0WWQ4 A0A1A8IIY3 A0A1A7YYD1 A0A1A8S5I9 A0A3Q0SBH2 A0A1E1X6C6 A0A2C9JGF0 A0A085NDM6 A0A3Q3A144 A0A3B3YCA9 A0A2U9BYK1 A0A3B3VDK4 A0A2I4C794 A0A3Q3MAN9 T1KEG9 A0A3P8YPV7 R7TQ29 A0A3Q1HIW5 A0A3Q3J341 A0A3Q1GMK9 A0A315WB05 A0A3B4CLM5 A0A0S7L5Q9 A0A3B5L107 A0A3P8SSQ0 A0A3Q2VXV4 A0A3Q1AUG3 A0A3P8Q9K3 A0A3P9D223 A0A3B5QE97 W4ZCP1 A0A3Q1GX48 A0A293LFE6 I3JHM1 A0A3Q3XGY2 A0A3B4G1L7 A0A3P9P2H6 A0A3B4UP26 A0A3B4YN92
A0A0N1PG54 A0A2J7Q743 A0A067R2N7 A0A2P8Y3F0 A0A026VZW6 A0A2A3EJ94 A0A087ZQ06 A0A151JU34 A0A232F8E1 A0A151IHN0 A0A151I5C9 K7ITZ8 A0A158NWS6 F4WQ34 A0A0M9A5L9 A0A154PFM3 E9IVZ5 A0A0L7QUQ7 A0A195EDC8 A0A310SUS1 A0A1W4XC59 A0A151WY70 A0A1Y1M253 A0A411G7L6 A0A0T6B631 A0A1S3H2L3 D2A5G5 T1J910 A0A087TK97 A0A2H4KB79 V5HD17 A0A131XZC1 A0A293LVL2 A0A2R5LM91 E0VK28 A0A224YVM6 B7QHR9 L7M8B3 A0A131YM47 A0A131XHY6 A0A0P4WT28 G3MPG1 A0A1E1XSV8 A0A210Q877 A0A443R7J4 V4AF39 A0A1A8EX67 A0A1A8CG66 A0A1A8DM65 A0A1A8MRD3 A0A1D1V3R2 A0A1A8UPU1 A0A1W0WWQ4 A0A1A8IIY3 A0A1A7YYD1 A0A1A8S5I9 A0A3Q0SBH2 A0A1E1X6C6 A0A2C9JGF0 A0A085NDM6 A0A3Q3A144 A0A3B3YCA9 A0A2U9BYK1 A0A3B3VDK4 A0A2I4C794 A0A3Q3MAN9 T1KEG9 A0A3P8YPV7 R7TQ29 A0A3Q1HIW5 A0A3Q3J341 A0A3Q1GMK9 A0A315WB05 A0A3B4CLM5 A0A0S7L5Q9 A0A3B5L107 A0A3P8SSQ0 A0A3Q2VXV4 A0A3Q1AUG3 A0A3P8Q9K3 A0A3P9D223 A0A3B5QE97 W4ZCP1 A0A3Q1GX48 A0A293LFE6 I3JHM1 A0A3Q3XGY2 A0A3B4G1L7 A0A3P9P2H6 A0A3B4UP26 A0A3B4YN92
PDB
3L4G
E-value=4.13099e-23,
Score=269
Ontologies
GO
Topology
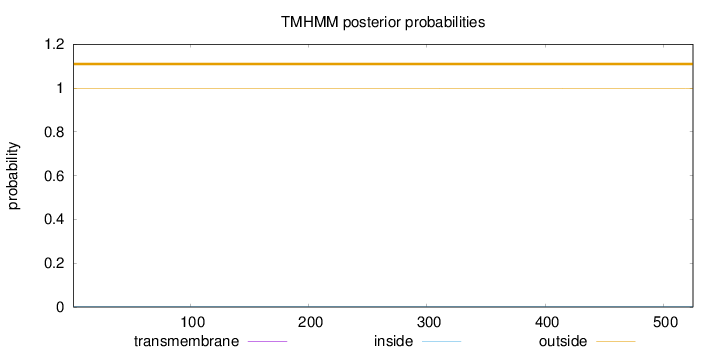
Length:
525
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00285
Exp number, first 60 AAs:
0.00102
Total prob of N-in:
0.00261
outside
1 - 525
Population Genetic Test Statistics
Pi
214.12063
Theta
172.591909
Tajima's D
0.406883
CLR
185.00319
CSRT
0.490225488725564
Interpretation
Uncertain
