Pre Gene Modal
BGIBMGA013073
Annotation
PREDICTED:_DNA_repair_and_recombination_protein_RAD54-like_[Amyelois_transitella]
Full name
DNA repair and recombination protein RAD54-like
Alternative Name
Protein okra
Location in the cell
Nuclear Reliability : 4.232
Sequence
CDS
ATGCGCCGCAGTCGAGCGCCAAGCCAAATAGGCTCTGGAGAGAGTGTTTTCAAGAGTCCATTACTAGACTCCGCCAAGCGAAAGAAACGAGAATGCACAAGAATACCGCTAGCACAGAAAACTCCTTCTCAAACTTTACCTTCTTCAGATCACGAAAACATTATCCGTCAAATACTTAGTAAGCCCTTTAAAGTTCCTATATCAAACTATGTCTCCTCGTATTGCAGTAAAAGCTTAGGATTAAAACGAACGCAACTTAGAAGGGCCCTCCACGACCCAGACGAACCCAATTCACTGGTCCTCGAACGCCCTCCAACCATTCCGGAACACGAAAGAATGAAAATGAATCCTAATGACATAAAAGTTGCTGTTGTAGTTGATCCAGTACTTGGAAACATACTGAGGCCGCACCAGAGAGAAGGAGTAAAGTTCATGTATGACTGCGTTACTGGGAAACAAATAGAAAATGCATATGGATGTATAATGGCTGATGAAATGGGTTTAGGTAAAACCTTGCAGTGCATAACTTTACTGTGGACCTTACTGCGTCAAGGGCCAGACTGTAAACCAACAATATGTAAAGCCATAATAGTATGTCCCAGCAGTTTAGTAAAAAATTGGTACAATGAAATACATAAGTGGCTGTCGCAAAGGATAAATGCTTTAGCTATGGATGGGGGTTCAAAAGCTGAAGTAACTTTGAAACTACAGCAATTTATGAACACATATTCTGCGACTCGAGTTGCCACACCAGTCTTAATCATATCATATGAAACTTTCAGGATATACTCAAATATATTGCACTCTTCTGAAGTAGGTTTAGTCCTGTGTGATGAAGGGCATAGGTTGAAGAACAGCGAAAATCAGACATACCAAGCATTAATGGGTTTGAAGGCTAAAAGGAGAATATTAATCTCAGGTACACCAATCCAGAATGATTTAACTGAATATTTTAGTTTAGTACATTTTGTTAATGAAGGAATATTAGGGACTGCGCATGAGTTCAAAAAGAGATATGAAAATCCTATTTTGAAGGGACAAGATGCAATGGCAAGTCAACCTGAGAGAGAAAAAGCCCAGGAGTGCCTGCAAACATTAACTTCAATTGTGAACAAGTGCATGATCCGAAGAACTAGTGGTCTATTAACTAAATATTTGCCAGTTAAATTTGAGCAAGTTATTTGTGTAAAAATGACTCCACTACAGACTCAAATTTACAAGAATTTCATAAATTCTGATGCAATTCGTAACAAGTTTACAGGCACTGGAGAAAAAAGCAATACGCTAAGTGCTTTGTCCAGCATTACATCTTTGAAGAAGCTCTGTAACCACCCAGACTTGATCTATGAAAAGATTATAGAAAGAACAGAAGGTTTCGAAAAAGCAAAAGACTATCTCCCAAGTAACTATGATATAAAAGATGTAAGACCAGAATATTCAGGCAAGTTAATGATCTTAGACTGCATACTGGCAAATCTTAAAATGAATACAGATGATAAAATTGTGCTCGTATCAAATTATACTCAGACATTAGATTTATTTGAGAAATTATGTCGAAAACGTACATATCAATATGTAAGATTGGATGGTTCAATGACAATAAAAAAGAGAGCCAAGGTGGTGGAAAACTTTAACAATAAAGACTCCAAAGAGTGGATTTTCATGCTAAGTTCCAAAGCTGGTGGTTGTGGCTTGAATCTCATTGGAGCCAACAGGCTAATCATGTTTGACCCTGATTGGAATCCTGCAAATGATGAGCAGGCGATGGCACGGGTATGGCGCGATGGGCAAAAAAAACCATGCTATATTTACAGACTTTTGGCTACTGGAACCATTGAAGAAAAAATATTCCAAAGGCAAGCACACAAAAAAGCACTCAGCGAGACAGTGGTTGACCAAAATGAGGAGTCTCTAAGACATTTCACAGCAGATGATTTGAAAGACCTATTCCGTTTGGAGGAGAACACACTATCGGACACTCATAGTAAGTTTAAATGTAAAAGGTGTGTAAACAATATTCAAGTTACACTACCACCTGAAAATTCTGATTGCACATCTGATATTTCCAATTGGTACCACTGTGCAGACAAGAAAAACATAGCCGACATGGTCTTGAAGCAATGCTGGGACGTTGCAAAAAGTATATCCTTTGTTTTCCATCATCGGTCCGCAAAAACAGAGGCTAAGGAGATAACTATAGAAGATAAAGAAAATATACAAGTAAGTAAAAAAAGAAGAATCATGGATGATGAAGATTACTCTGAACCAGATGACAGTGCTGATGAAGATTATTCATGA
Protein
MRRSRAPSQIGSGESVFKSPLLDSAKRKKRECTRIPLAQKTPSQTLPSSDHENIIRQILSKPFKVPISNYVSSYCSKSLGLKRTQLRRALHDPDEPNSLVLERPPTIPEHERMKMNPNDIKVAVVVDPVLGNILRPHQREGVKFMYDCVTGKQIENAYGCIMADEMGLGKTLQCITLLWTLLRQGPDCKPTICKAIIVCPSSLVKNWYNEIHKWLSQRINALAMDGGSKAEVTLKLQQFMNTYSATRVATPVLIISYETFRIYSNILHSSEVGLVLCDEGHRLKNSENQTYQALMGLKAKRRILISGTPIQNDLTEYFSLVHFVNEGILGTAHEFKKRYENPILKGQDAMASQPEREKAQECLQTLTSIVNKCMIRRTSGLLTKYLPVKFEQVICVKMTPLQTQIYKNFINSDAIRNKFTGTGEKSNTLSALSSITSLKKLCNHPDLIYEKIIERTEGFEKAKDYLPSNYDIKDVRPEYSGKLMILDCILANLKMNTDDKIVLVSNYTQTLDLFEKLCRKRTYQYVRLDGSMTIKKRAKVVENFNNKDSKEWIFMLSSKAGGCGLNLIGANRLIMFDPDWNPANDEQAMARVWRDGQKKPCYIYRLLATGTIEEKIFQRQAHKKALSETVVDQNEESLRHFTADDLKDLFRLEENTLSDTHSKFKCKRCVNNIQVTLPPENSDCTSDISNWYHCADKKNIADMVLKQCWDVAKSISFVFHHRSAKTEAKEITIEDKENIQVSKKRRIMDDEDYSEPDDSADEDYS
Summary
Description
Involved in mitotic DNA repair and meiotic recombination. Functions in the recombinational DNA repair pathway. Essential for interhomolog gene conversion (GC), but may have a less important role in intersister GC than spn-A/Rad51. In the presence of DNA, spn-A/Rad51 enhances the ATPase activity of okr/Rad54 (By similarity).
Subunit
Interacts (via N-terminus) with spn-A/Rad51.
Similarity
Belongs to the SNF2/RAD54 helicase family.
Keywords
ATP-binding
Cell cycle
Cell division
Complete proteome
DNA damage
DNA repair
DNA-binding
Helicase
Hydrolase
Meiosis
Mitosis
Nucleotide-binding
Nucleus
Phosphoprotein
Reference proteome
Feature
chain DNA repair and recombination protein RAD54-like
Uniprot
H9JU61
A0A437BBY4
A0A2A4K3C5
A0A0N0PC66
A0A194Q0D7
A0A0N0P9X5
+ More
A0A2H1VXF0 A0A212FMW8 A0A0L7LE18 Q17HL0 B0WI46 A0A1Q3F704 A0A1W4WLL8 A0A1B0C8Y4 A0A182JLX7 A0A336M061 A0A182MGC7 A0A182Q2T2 A0A182VTJ4 A0A1S4GZV0 A0A182K5X1 A0A182KWJ7 A0A182PVT7 A0A182R433 A0A182VJ32 A0A182TNY4 Q5TS22 A0A182IB86 A0A182NB95 A0A182F3P9 A0A182X665 K7J2D0 A0A232EWT2 W5JSB4 A0A026X3E4 E0VYD7 A0A291S6X6 A0A158NVK9 A0A195BMY9 A0A067R8G3 F4WLD6 A0A0C9RBG6 A0A151IZS8 E2AP89 D2A5V0 A0A154PMN2 A0A0J7KYW1 A0A1Y1LRC4 A0A088ASJ5 A0A2J7QMJ3 A0A0L0BW48 A0A1I8MAA9 A0A0L7QQS0 A0A1A9V1B1 E2BNJ2 A0A0N0BK38 A0A1B6D4V7 A0A2A3EDH5 A0A0A1WVB9 A0A1B6J4K0 A0A3Q0J4X1 B4KHL5 A0A1B0A884 A0A1A9X858 A0A433TNT2 A0A1B0G048 A0A1Y1LL94 W8BQN5 A0A1I8NPK8 N6TY08 B4M9A8 A0A0K8W604 A0A034WNT0 A0A1S3IIK1 B4MX21 A0A0M3QT64 A0A3N0Y5Z8 B4JCS7 A0A1J1IKR6 A0A0P5E4U1 A0A0P5ZJK8 A0A0P5V8H7 A0A0P6GD54 A0A0P5SNY4 A0A0P5INJ7 A0A0P5HUV8 A0A0P5UUM8 A0A0N8AV34 A0A0P5GT85 A0A0N8D808 A0A0P6G8V2 A0A146P5K0 A0A3P9Q5M1 V4A080 A0A401S9R4 A0A2Y9ITX4 E9GW91 A0A3Q7RQ66 W4XZN5 F1PJC9
A0A2H1VXF0 A0A212FMW8 A0A0L7LE18 Q17HL0 B0WI46 A0A1Q3F704 A0A1W4WLL8 A0A1B0C8Y4 A0A182JLX7 A0A336M061 A0A182MGC7 A0A182Q2T2 A0A182VTJ4 A0A1S4GZV0 A0A182K5X1 A0A182KWJ7 A0A182PVT7 A0A182R433 A0A182VJ32 A0A182TNY4 Q5TS22 A0A182IB86 A0A182NB95 A0A182F3P9 A0A182X665 K7J2D0 A0A232EWT2 W5JSB4 A0A026X3E4 E0VYD7 A0A291S6X6 A0A158NVK9 A0A195BMY9 A0A067R8G3 F4WLD6 A0A0C9RBG6 A0A151IZS8 E2AP89 D2A5V0 A0A154PMN2 A0A0J7KYW1 A0A1Y1LRC4 A0A088ASJ5 A0A2J7QMJ3 A0A0L0BW48 A0A1I8MAA9 A0A0L7QQS0 A0A1A9V1B1 E2BNJ2 A0A0N0BK38 A0A1B6D4V7 A0A2A3EDH5 A0A0A1WVB9 A0A1B6J4K0 A0A3Q0J4X1 B4KHL5 A0A1B0A884 A0A1A9X858 A0A433TNT2 A0A1B0G048 A0A1Y1LL94 W8BQN5 A0A1I8NPK8 N6TY08 B4M9A8 A0A0K8W604 A0A034WNT0 A0A1S3IIK1 B4MX21 A0A0M3QT64 A0A3N0Y5Z8 B4JCS7 A0A1J1IKR6 A0A0P5E4U1 A0A0P5ZJK8 A0A0P5V8H7 A0A0P6GD54 A0A0P5SNY4 A0A0P5INJ7 A0A0P5HUV8 A0A0P5UUM8 A0A0N8AV34 A0A0P5GT85 A0A0N8D808 A0A0P6G8V2 A0A146P5K0 A0A3P9Q5M1 V4A080 A0A401S9R4 A0A2Y9ITX4 E9GW91 A0A3Q7RQ66 W4XZN5 F1PJC9
EC Number
3.6.4.-
Pubmed
19121390
26354079
22118469
26227816
17510324
12364791
+ More
20966253 20075255 28648823 20920257 23761445 24508170 30249741 20566863 28992199 21347285 24845553 21719571 20798317 18362917 19820115 28004739 26108605 25315136 25830018 17994087 24495485 23537049 25348373 23254933 30297745 21292972 16341006
20966253 20075255 28648823 20920257 23761445 24508170 30249741 20566863 28992199 21347285 24845553 21719571 20798317 18362917 19820115 28004739 26108605 25315136 25830018 17994087 24495485 23537049 25348373 23254933 30297745 21292972 16341006
EMBL
BABH01004144
RSAL01000093
RVE47910.1
NWSH01000207
PCG78404.1
KQ460651
+ More
KPJ13057.1 KQ459582 KPI99021.1 KQ459058 KPJ03868.1 ODYU01005031 SOQ45510.1 AGBW02007651 OWR55094.1 JTDY01001478 KOB73773.1 CH477247 EAT46147.1 DS231942 EDS28209.1 GFDL01011742 JAV23303.1 AJWK01001599 UFQS01000376 UFQT01000376 SSX03350.1 SSX23716.1 AXCM01003320 AXCN02000632 AAAB01008944 EAL40287.3 APCN01000649 AAZX01001562 NNAY01001822 OXU22814.1 ADMH02000240 ETN67272.1 KK107019 QOIP01000007 EZA62787.1 RLU20477.1 DS235845 EEB18393.1 MF433009 ATL75360.1 ADTU01027231 KQ976433 KYM87345.1 KK852812 KDR15866.1 GL888207 EGI65099.1 GBYB01006780 GBYB01006781 GBYB01013834 GBYB01013835 JAG76547.1 JAG76548.1 JAG83601.1 JAG83602.1 KQ980673 KYN14605.1 GL441481 EFN64762.1 KQ971345 EFA05805.2 KQ434948 KZC12470.1 LBMM01001801 KMQ95722.1 GEZM01052529 JAV74455.1 NEVH01013208 PNF29788.1 JRES01001249 KNC24241.1 KQ414785 KOC60978.1 GL449414 EFN82771.1 KQ435710 KOX79922.1 GEDC01016556 JAS20742.1 KZ288271 PBC29777.1 GBXI01011959 JAD02333.1 GECU01013619 JAS94087.1 CH933807 EDW12294.1 RQTK01000254 RUS83190.1 CCAG010002366 GEZM01052530 JAV74454.1 GAMC01010999 JAB95556.1 APGK01050691 APGK01050692 APGK01050693 KB741174 KB631692 ENN73266.1 ERL85409.1 CH940654 EDW57784.1 GDHF01005830 JAI46484.1 GAKP01002945 JAC56007.1 CH963857 EDW76660.1 CP012523 ALC38339.1 RJVU01051519 ROL41636.1 CH916368 EDW03166.1 CVRI01000054 CRL00768.1 GDIP01146531 JAJ76871.1 GDIP01046289 JAM57426.1 GDIP01103274 JAM00441.1 GDIQ01035023 JAN59714.1 GDIP01137342 JAL66372.1 GDIQ01212732 JAK38993.1 GDIQ01245429 JAK06296.1 GDIP01108482 JAL95232.1 GDIQ01239987 JAK11738.1 GDIQ01237185 JAK14540.1 GDIP01059783 JAM43932.1 GDIQ01038465 JAN56272.1 GCES01147081 JAQ39241.1 KB203019 ESO86661.1 BEZZ01000151 GCC27090.1 GL732569 EFX76308.1 AAGJ04156460 AAGJ04156461 AAGJ04156462 AAEX03009776
KPJ13057.1 KQ459582 KPI99021.1 KQ459058 KPJ03868.1 ODYU01005031 SOQ45510.1 AGBW02007651 OWR55094.1 JTDY01001478 KOB73773.1 CH477247 EAT46147.1 DS231942 EDS28209.1 GFDL01011742 JAV23303.1 AJWK01001599 UFQS01000376 UFQT01000376 SSX03350.1 SSX23716.1 AXCM01003320 AXCN02000632 AAAB01008944 EAL40287.3 APCN01000649 AAZX01001562 NNAY01001822 OXU22814.1 ADMH02000240 ETN67272.1 KK107019 QOIP01000007 EZA62787.1 RLU20477.1 DS235845 EEB18393.1 MF433009 ATL75360.1 ADTU01027231 KQ976433 KYM87345.1 KK852812 KDR15866.1 GL888207 EGI65099.1 GBYB01006780 GBYB01006781 GBYB01013834 GBYB01013835 JAG76547.1 JAG76548.1 JAG83601.1 JAG83602.1 KQ980673 KYN14605.1 GL441481 EFN64762.1 KQ971345 EFA05805.2 KQ434948 KZC12470.1 LBMM01001801 KMQ95722.1 GEZM01052529 JAV74455.1 NEVH01013208 PNF29788.1 JRES01001249 KNC24241.1 KQ414785 KOC60978.1 GL449414 EFN82771.1 KQ435710 KOX79922.1 GEDC01016556 JAS20742.1 KZ288271 PBC29777.1 GBXI01011959 JAD02333.1 GECU01013619 JAS94087.1 CH933807 EDW12294.1 RQTK01000254 RUS83190.1 CCAG010002366 GEZM01052530 JAV74454.1 GAMC01010999 JAB95556.1 APGK01050691 APGK01050692 APGK01050693 KB741174 KB631692 ENN73266.1 ERL85409.1 CH940654 EDW57784.1 GDHF01005830 JAI46484.1 GAKP01002945 JAC56007.1 CH963857 EDW76660.1 CP012523 ALC38339.1 RJVU01051519 ROL41636.1 CH916368 EDW03166.1 CVRI01000054 CRL00768.1 GDIP01146531 JAJ76871.1 GDIP01046289 JAM57426.1 GDIP01103274 JAM00441.1 GDIQ01035023 JAN59714.1 GDIP01137342 JAL66372.1 GDIQ01212732 JAK38993.1 GDIQ01245429 JAK06296.1 GDIP01108482 JAL95232.1 GDIQ01239987 JAK11738.1 GDIQ01237185 JAK14540.1 GDIP01059783 JAM43932.1 GDIQ01038465 JAN56272.1 GCES01147081 JAQ39241.1 KB203019 ESO86661.1 BEZZ01000151 GCC27090.1 GL732569 EFX76308.1 AAGJ04156460 AAGJ04156461 AAGJ04156462 AAEX03009776
Proteomes
UP000005204
UP000283053
UP000218220
UP000053240
UP000053268
UP000007151
+ More
UP000037510 UP000008820 UP000002320 UP000192223 UP000092461 UP000075880 UP000075883 UP000075886 UP000075920 UP000075881 UP000075882 UP000075885 UP000075900 UP000075903 UP000075902 UP000007062 UP000075840 UP000075884 UP000069272 UP000076407 UP000002358 UP000215335 UP000000673 UP000053097 UP000279307 UP000009046 UP000005205 UP000078540 UP000027135 UP000007755 UP000078492 UP000000311 UP000007266 UP000076502 UP000036403 UP000005203 UP000235965 UP000037069 UP000095301 UP000053825 UP000078200 UP000008237 UP000053105 UP000242457 UP000079169 UP000009192 UP000092445 UP000092443 UP000271974 UP000092444 UP000095300 UP000019118 UP000030742 UP000008792 UP000085678 UP000007798 UP000092553 UP000001070 UP000183832 UP000242638 UP000030746 UP000287033 UP000248482 UP000000305 UP000286640 UP000007110 UP000002254
UP000037510 UP000008820 UP000002320 UP000192223 UP000092461 UP000075880 UP000075883 UP000075886 UP000075920 UP000075881 UP000075882 UP000075885 UP000075900 UP000075903 UP000075902 UP000007062 UP000075840 UP000075884 UP000069272 UP000076407 UP000002358 UP000215335 UP000000673 UP000053097 UP000279307 UP000009046 UP000005205 UP000078540 UP000027135 UP000007755 UP000078492 UP000000311 UP000007266 UP000076502 UP000036403 UP000005203 UP000235965 UP000037069 UP000095301 UP000053825 UP000078200 UP000008237 UP000053105 UP000242457 UP000079169 UP000009192 UP000092445 UP000092443 UP000271974 UP000092444 UP000095300 UP000019118 UP000030742 UP000008792 UP000085678 UP000007798 UP000092553 UP000001070 UP000183832 UP000242638 UP000030746 UP000287033 UP000248482 UP000000305 UP000286640 UP000007110 UP000002254
Interpro
IPR038718
SNF2-like_sf
+ More
IPR027417 P-loop_NTPase
IPR000330 SNF2_N
IPR001650 Helicase_C
IPR014001 Helicase_ATP-bd
IPR032675 LRR_dom_sf
IPR005146 B3/B4_tRNA-bd
IPR020825 Phe-tRNA_synthase_B3/B4
IPR001611 Leu-rich_rpt
IPR003591 Leu-rich_rpt_typical-subtyp
IPR013967 Rad54_N
IPR036855 Znf_CCCH_sf
IPR000571 Znf_CCCH
IPR027417 P-loop_NTPase
IPR000330 SNF2_N
IPR001650 Helicase_C
IPR014001 Helicase_ATP-bd
IPR032675 LRR_dom_sf
IPR005146 B3/B4_tRNA-bd
IPR020825 Phe-tRNA_synthase_B3/B4
IPR001611 Leu-rich_rpt
IPR003591 Leu-rich_rpt_typical-subtyp
IPR013967 Rad54_N
IPR036855 Znf_CCCH_sf
IPR000571 Znf_CCCH
Gene 3D
CDD
ProteinModelPortal
H9JU61
A0A437BBY4
A0A2A4K3C5
A0A0N0PC66
A0A194Q0D7
A0A0N0P9X5
+ More
A0A2H1VXF0 A0A212FMW8 A0A0L7LE18 Q17HL0 B0WI46 A0A1Q3F704 A0A1W4WLL8 A0A1B0C8Y4 A0A182JLX7 A0A336M061 A0A182MGC7 A0A182Q2T2 A0A182VTJ4 A0A1S4GZV0 A0A182K5X1 A0A182KWJ7 A0A182PVT7 A0A182R433 A0A182VJ32 A0A182TNY4 Q5TS22 A0A182IB86 A0A182NB95 A0A182F3P9 A0A182X665 K7J2D0 A0A232EWT2 W5JSB4 A0A026X3E4 E0VYD7 A0A291S6X6 A0A158NVK9 A0A195BMY9 A0A067R8G3 F4WLD6 A0A0C9RBG6 A0A151IZS8 E2AP89 D2A5V0 A0A154PMN2 A0A0J7KYW1 A0A1Y1LRC4 A0A088ASJ5 A0A2J7QMJ3 A0A0L0BW48 A0A1I8MAA9 A0A0L7QQS0 A0A1A9V1B1 E2BNJ2 A0A0N0BK38 A0A1B6D4V7 A0A2A3EDH5 A0A0A1WVB9 A0A1B6J4K0 A0A3Q0J4X1 B4KHL5 A0A1B0A884 A0A1A9X858 A0A433TNT2 A0A1B0G048 A0A1Y1LL94 W8BQN5 A0A1I8NPK8 N6TY08 B4M9A8 A0A0K8W604 A0A034WNT0 A0A1S3IIK1 B4MX21 A0A0M3QT64 A0A3N0Y5Z8 B4JCS7 A0A1J1IKR6 A0A0P5E4U1 A0A0P5ZJK8 A0A0P5V8H7 A0A0P6GD54 A0A0P5SNY4 A0A0P5INJ7 A0A0P5HUV8 A0A0P5UUM8 A0A0N8AV34 A0A0P5GT85 A0A0N8D808 A0A0P6G8V2 A0A146P5K0 A0A3P9Q5M1 V4A080 A0A401S9R4 A0A2Y9ITX4 E9GW91 A0A3Q7RQ66 W4XZN5 F1PJC9
A0A2H1VXF0 A0A212FMW8 A0A0L7LE18 Q17HL0 B0WI46 A0A1Q3F704 A0A1W4WLL8 A0A1B0C8Y4 A0A182JLX7 A0A336M061 A0A182MGC7 A0A182Q2T2 A0A182VTJ4 A0A1S4GZV0 A0A182K5X1 A0A182KWJ7 A0A182PVT7 A0A182R433 A0A182VJ32 A0A182TNY4 Q5TS22 A0A182IB86 A0A182NB95 A0A182F3P9 A0A182X665 K7J2D0 A0A232EWT2 W5JSB4 A0A026X3E4 E0VYD7 A0A291S6X6 A0A158NVK9 A0A195BMY9 A0A067R8G3 F4WLD6 A0A0C9RBG6 A0A151IZS8 E2AP89 D2A5V0 A0A154PMN2 A0A0J7KYW1 A0A1Y1LRC4 A0A088ASJ5 A0A2J7QMJ3 A0A0L0BW48 A0A1I8MAA9 A0A0L7QQS0 A0A1A9V1B1 E2BNJ2 A0A0N0BK38 A0A1B6D4V7 A0A2A3EDH5 A0A0A1WVB9 A0A1B6J4K0 A0A3Q0J4X1 B4KHL5 A0A1B0A884 A0A1A9X858 A0A433TNT2 A0A1B0G048 A0A1Y1LL94 W8BQN5 A0A1I8NPK8 N6TY08 B4M9A8 A0A0K8W604 A0A034WNT0 A0A1S3IIK1 B4MX21 A0A0M3QT64 A0A3N0Y5Z8 B4JCS7 A0A1J1IKR6 A0A0P5E4U1 A0A0P5ZJK8 A0A0P5V8H7 A0A0P6GD54 A0A0P5SNY4 A0A0P5INJ7 A0A0P5HUV8 A0A0P5UUM8 A0A0N8AV34 A0A0P5GT85 A0A0N8D808 A0A0P6G8V2 A0A146P5K0 A0A3P9Q5M1 V4A080 A0A401S9R4 A0A2Y9ITX4 E9GW91 A0A3Q7RQ66 W4XZN5 F1PJC9
PDB
1Z3I
E-value=0,
Score=2247
Ontologies
GO
GO:0005524
GO:0003723
GO:0004826
GO:0016817
GO:0046872
GO:0005634
GO:0010212
GO:0003677
GO:0000711
GO:0006338
GO:0004386
GO:0051301
GO:0043150
GO:0000724
GO:0030716
GO:0048477
GO:0007131
GO:0045003
GO:0042493
GO:0008340
GO:0032991
GO:0036310
GO:0005654
GO:0005737
GO:0008299
GO:0008152
GO:0016301
GO:0016773
GO:0006418
GO:0000049
GO:0016876
Topology
Subcellular location
Nucleus
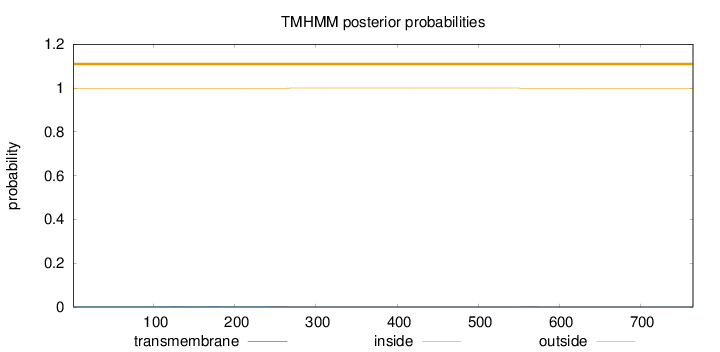
Length:
765
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02882
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00144
outside
1 - 765
Population Genetic Test Statistics
Pi
242.098223
Theta
202.882808
Tajima's D
-0.677826
CLR
26.615963
CSRT
0.2012399380031
Interpretation
Uncertain
