Gene
KWMTBOMO09449
Pre Gene Modal
BGIBMGA012785
Annotation
PREDICTED:_selenoprotein_O-like_[Plutella_xylostella]
Full name
Protein adenylyltransferase SelO
Location in the cell
Cytoplasmic Reliability : 1.418 Extracellular Reliability : 1.137 Nuclear Reliability : 1.018
Sequence
CDS
ATGAAGATCGGTTTGAAAGAAGAGCGTTGGGGAGACGAGCAGCTGGTAGAGAAGTTGTTAGACATGATGCAGCAGACTGGAGCCGATTTTACTGCTACGTTCAGGCAACTAGCTGAGTTGGAACCATCGGAGCTGGTCAGCGAAGTGAAGATATCAGAGAAGTGGTCGCTCAAACGTTTGTCGTCTCACGCTTCGTGGGGCTGCTGGCTCGACCAGTATCGTGAGAGGCTCGACAAGGAAGCTGGTGGGTTTTAA
Protein
MKIGLKEERWGDEQLVEKLLDMMQQTGADFTATFRQLAELEPSELVSEVKISEKWSLKRLSSHASWGCWLDQYRERLDKEAGGF
Summary
Description
Catalyzes the transfer of adenosine 5'-monophosphate (AMP) to Ser, Thr or Tyr residues of target proteins (AMPylation).
Catalytic Activity
ATP + L-seryl-[protein] = 3-O-(5'-adenylyl)-L-seryl-[protein] + diphosphate
ATP + L-threonyl-[protein] = 3-O-(5'-adenylyl)-L-threonyl-[protein] + diphosphate
ATP + L-tyrosyl-[protein] = diphosphate + O-(5'-adenylyl)-L-tyrosyl-[protein]
ATP + L-threonyl-[protein] = 3-O-(5'-adenylyl)-L-threonyl-[protein] + diphosphate
ATP + L-tyrosyl-[protein] = diphosphate + O-(5'-adenylyl)-L-tyrosyl-[protein]
Cofactor
Mg(2+)
Similarity
Belongs to the SELO family.
Uniprot
H9JTC3
A0A2A4K3S9
A0A2H1V3U7
A0A3S2LZX5
A0A194Q0B0
A0A194R1J0
+ More
A0A212FN18 A0A0L7LNE1 A0A3S2NCZ1 A0A2H1VXE5 A0A2A4K2D8 A0A194Q6J4 A0A212FMZ4 A0A2P8Z331 S4PZ13 A0A452FWM8 W5Q4A1 A0A1U7TG25 A0A3Q2HUH5 A0A1S3F2L4 A0A1S3F024 G1Q4G1 A0A384DMA8 A0A3Q7VTS7 A0A3Q7QMU4 A0A3Q7S2Y6 A0A3Q7V2D3 A0A384DM74 A0A3Q7UCR6 A0A384DLL3 A0A3Q7QQ14 A0A2Y9IHL2 A0A3Q7W6U9 A0A3Q7S2Y1 A0A2Y9IGM5 A0A2Y9INX6 A0A2Y9GUB2 A0A2Y9IHL7 A0A3Q7QPJ6 A0A2T3ED75 L8HXH5 A0A452FXE2 M3YLC1 A0A452FWN8 A0A2U3YQS3 A0A3Q1M8C2 A0A3P4M993 A0A1E4Y0N6 A0A0N1I3L0 A0A3Q1BVP7 A0A067QYY2 A0A3Q2Q7P6 A0A146ZRX7 A0A2U3W770 A0A146MT68 A0A146MRF5 A0A146MRI0 A0A248W3R1 A0A3M6U6Z3 M3WX42 A0A1W4YYD7 A0A2J7PTW5 A0A2J7PTW2 A0A2Y9NYC1 A0A3P8XS56 A0A2Y9P1S9 A0A2B4SDX5 A0A340YEQ1 A0A341B8R2 A0A452C6Z3 A0A2J7PTW3 A0A3Q2WUM8 I3JGT7 A0A2J7PTW6 A0A1C3V031 A0A210Q0I0 A0A401SVU4 A0A1H9V393 A0A1C3UG58 A0A3B4ZN00 B3S131 A0A0A8GDC9 A0A2A6JHF4 J0W035 A0A3B4ZL86 A0A369SLN6 A0A3Q3F0M6 A0A369C1E1 C5LW11 A0A0M3GLY8 A0A192T5J0 G3NN69 A0A444HVZ8 A0A1B9SDI1 A0A3Q3W811 A0A372K6B1 A0A3S3WY66 A0A329YPC5 A7SUL1 A0A2U0WLP7
A0A212FN18 A0A0L7LNE1 A0A3S2NCZ1 A0A2H1VXE5 A0A2A4K2D8 A0A194Q6J4 A0A212FMZ4 A0A2P8Z331 S4PZ13 A0A452FWM8 W5Q4A1 A0A1U7TG25 A0A3Q2HUH5 A0A1S3F2L4 A0A1S3F024 G1Q4G1 A0A384DMA8 A0A3Q7VTS7 A0A3Q7QMU4 A0A3Q7S2Y6 A0A3Q7V2D3 A0A384DM74 A0A3Q7UCR6 A0A384DLL3 A0A3Q7QQ14 A0A2Y9IHL2 A0A3Q7W6U9 A0A3Q7S2Y1 A0A2Y9IGM5 A0A2Y9INX6 A0A2Y9GUB2 A0A2Y9IHL7 A0A3Q7QPJ6 A0A2T3ED75 L8HXH5 A0A452FXE2 M3YLC1 A0A452FWN8 A0A2U3YQS3 A0A3Q1M8C2 A0A3P4M993 A0A1E4Y0N6 A0A0N1I3L0 A0A3Q1BVP7 A0A067QYY2 A0A3Q2Q7P6 A0A146ZRX7 A0A2U3W770 A0A146MT68 A0A146MRF5 A0A146MRI0 A0A248W3R1 A0A3M6U6Z3 M3WX42 A0A1W4YYD7 A0A2J7PTW5 A0A2J7PTW2 A0A2Y9NYC1 A0A3P8XS56 A0A2Y9P1S9 A0A2B4SDX5 A0A340YEQ1 A0A341B8R2 A0A452C6Z3 A0A2J7PTW3 A0A3Q2WUM8 I3JGT7 A0A2J7PTW6 A0A1C3V031 A0A210Q0I0 A0A401SVU4 A0A1H9V393 A0A1C3UG58 A0A3B4ZN00 B3S131 A0A0A8GDC9 A0A2A6JHF4 J0W035 A0A3B4ZL86 A0A369SLN6 A0A3Q3F0M6 A0A369C1E1 C5LW11 A0A0M3GLY8 A0A192T5J0 G3NN69 A0A444HVZ8 A0A1B9SDI1 A0A3Q3W811 A0A372K6B1 A0A3S3WY66 A0A329YPC5 A7SUL1 A0A2U0WLP7
EC Number
2.7.7.-
2.7.7.n1
2.7.7.n1
Pubmed
EMBL
BABH01004144
NWSH01000207
PCG78423.1
ODYU01000368
SOQ35032.1
RSAL01000093
+ More
RVE47912.1 KQ459582 KPI99017.1 KQ460878 KPJ11663.1 AGBW02007651 OWR55099.1 JTDY01000502 KOB76874.1 RVE47911.1 ODYU01005031 SOQ45509.1 PCG78425.1 KPI99020.1 OWR55093.1 PYGN01000219 PSN50911.1 GAIX01002338 JAA90222.1 LWLT01000014 AMGL01024142 AAPE02040420 AAPE02040421 AAPE02040422 AAPE02040423 PYJQ01000005 PST20906.1 JH882683 ELR48546.1 AEYP01043726 AEYP01043727 CYRY02008221 VCW76992.1 LZPI01000069 OEC92923.1 KQ459058 KPJ03869.1 KK853093 KDR11508.1 GCES01017254 JAR69069.1 GCES01164040 JAQ22282.1 GCES01164041 JAQ22281.1 GCES01164039 JAQ22283.1 CP022998 ASW05904.1 RCHS01002141 RMX49440.1 AANG04002256 NEVH01021221 PNF19773.1 PNF19770.1 LSMT01000106 PFX27299.1 PNF19772.1 AERX01003104 PNF19771.1 FMAG01000002 SCB21035.1 NEDP02005302 OWF42250.1 BEZZ01000605 GCC34501.1 FOGR01000003 SES16290.1 FMAC01000002 SCB14327.1 DS985247 EDV23495.1 CP007641 AJC78489.1 NWSV01000003 PDT05372.1 JH719395 EJC78468.1 NOWV01000001 RDD47790.1 QPJY01000009 RCX26487.1 GG686046 EEQ99081.1 AHJU02000012 KKZ87311.1 CP013574 ANL90553.1 SBHX01000049 RWX27983.1 LXKN01000001 OCJ11748.1 QVQV01000002 RFU47742.1 SBJG01000004 RWY80273.1 QMKK01000019 RAX43100.1 DS469814 EDO32626.1 QEOE01000014 PVX83061.1
RVE47912.1 KQ459582 KPI99017.1 KQ460878 KPJ11663.1 AGBW02007651 OWR55099.1 JTDY01000502 KOB76874.1 RVE47911.1 ODYU01005031 SOQ45509.1 PCG78425.1 KPI99020.1 OWR55093.1 PYGN01000219 PSN50911.1 GAIX01002338 JAA90222.1 LWLT01000014 AMGL01024142 AAPE02040420 AAPE02040421 AAPE02040422 AAPE02040423 PYJQ01000005 PST20906.1 JH882683 ELR48546.1 AEYP01043726 AEYP01043727 CYRY02008221 VCW76992.1 LZPI01000069 OEC92923.1 KQ459058 KPJ03869.1 KK853093 KDR11508.1 GCES01017254 JAR69069.1 GCES01164040 JAQ22282.1 GCES01164041 JAQ22281.1 GCES01164039 JAQ22283.1 CP022998 ASW05904.1 RCHS01002141 RMX49440.1 AANG04002256 NEVH01021221 PNF19773.1 PNF19770.1 LSMT01000106 PFX27299.1 PNF19772.1 AERX01003104 PNF19771.1 FMAG01000002 SCB21035.1 NEDP02005302 OWF42250.1 BEZZ01000605 GCC34501.1 FOGR01000003 SES16290.1 FMAC01000002 SCB14327.1 DS985247 EDV23495.1 CP007641 AJC78489.1 NWSV01000003 PDT05372.1 JH719395 EJC78468.1 NOWV01000001 RDD47790.1 QPJY01000009 RCX26487.1 GG686046 EEQ99081.1 AHJU02000012 KKZ87311.1 CP013574 ANL90553.1 SBHX01000049 RWX27983.1 LXKN01000001 OCJ11748.1 QVQV01000002 RFU47742.1 SBJG01000004 RWY80273.1 QMKK01000019 RAX43100.1 DS469814 EDO32626.1 QEOE01000014 PVX83061.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000053240
UP000007151
+ More
UP000037510 UP000245037 UP000291000 UP000002356 UP000189704 UP000002281 UP000081671 UP000001074 UP000261680 UP000286642 UP000286641 UP000291021 UP000248482 UP000248481 UP000243756 UP000000715 UP000245341 UP000009136 UP000095065 UP000257160 UP000027135 UP000265000 UP000245340 UP000214945 UP000275408 UP000011712 UP000192224 UP000235965 UP000248483 UP000265140 UP000225706 UP000265300 UP000252040 UP000261681 UP000264840 UP000005207 UP000199101 UP000242188 UP000287033 UP000198532 UP000186228 UP000261400 UP000009022 UP000031160 UP000220768 UP000005732 UP000253843 UP000261660 UP000252707 UP000034829 UP000078566 UP000007635 UP000283817 UP000093320 UP000261620 UP000263097 UP000287619 UP000251205 UP000001593 UP000245797
UP000037510 UP000245037 UP000291000 UP000002356 UP000189704 UP000002281 UP000081671 UP000001074 UP000261680 UP000286642 UP000286641 UP000291021 UP000248482 UP000248481 UP000243756 UP000000715 UP000245341 UP000009136 UP000095065 UP000257160 UP000027135 UP000265000 UP000245340 UP000214945 UP000275408 UP000011712 UP000192224 UP000235965 UP000248483 UP000265140 UP000225706 UP000265300 UP000252040 UP000261681 UP000264840 UP000005207 UP000199101 UP000242188 UP000287033 UP000198532 UP000186228 UP000261400 UP000009022 UP000031160 UP000220768 UP000005732 UP000253843 UP000261660 UP000252707 UP000034829 UP000078566 UP000007635 UP000283817 UP000093320 UP000261620 UP000263097 UP000287619 UP000251205 UP000001593 UP000245797
PRIDE
Pfam
PF02696 UPF0061
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
H9JTC3
A0A2A4K3S9
A0A2H1V3U7
A0A3S2LZX5
A0A194Q0B0
A0A194R1J0
+ More
A0A212FN18 A0A0L7LNE1 A0A3S2NCZ1 A0A2H1VXE5 A0A2A4K2D8 A0A194Q6J4 A0A212FMZ4 A0A2P8Z331 S4PZ13 A0A452FWM8 W5Q4A1 A0A1U7TG25 A0A3Q2HUH5 A0A1S3F2L4 A0A1S3F024 G1Q4G1 A0A384DMA8 A0A3Q7VTS7 A0A3Q7QMU4 A0A3Q7S2Y6 A0A3Q7V2D3 A0A384DM74 A0A3Q7UCR6 A0A384DLL3 A0A3Q7QQ14 A0A2Y9IHL2 A0A3Q7W6U9 A0A3Q7S2Y1 A0A2Y9IGM5 A0A2Y9INX6 A0A2Y9GUB2 A0A2Y9IHL7 A0A3Q7QPJ6 A0A2T3ED75 L8HXH5 A0A452FXE2 M3YLC1 A0A452FWN8 A0A2U3YQS3 A0A3Q1M8C2 A0A3P4M993 A0A1E4Y0N6 A0A0N1I3L0 A0A3Q1BVP7 A0A067QYY2 A0A3Q2Q7P6 A0A146ZRX7 A0A2U3W770 A0A146MT68 A0A146MRF5 A0A146MRI0 A0A248W3R1 A0A3M6U6Z3 M3WX42 A0A1W4YYD7 A0A2J7PTW5 A0A2J7PTW2 A0A2Y9NYC1 A0A3P8XS56 A0A2Y9P1S9 A0A2B4SDX5 A0A340YEQ1 A0A341B8R2 A0A452C6Z3 A0A2J7PTW3 A0A3Q2WUM8 I3JGT7 A0A2J7PTW6 A0A1C3V031 A0A210Q0I0 A0A401SVU4 A0A1H9V393 A0A1C3UG58 A0A3B4ZN00 B3S131 A0A0A8GDC9 A0A2A6JHF4 J0W035 A0A3B4ZL86 A0A369SLN6 A0A3Q3F0M6 A0A369C1E1 C5LW11 A0A0M3GLY8 A0A192T5J0 G3NN69 A0A444HVZ8 A0A1B9SDI1 A0A3Q3W811 A0A372K6B1 A0A3S3WY66 A0A329YPC5 A7SUL1 A0A2U0WLP7
A0A212FN18 A0A0L7LNE1 A0A3S2NCZ1 A0A2H1VXE5 A0A2A4K2D8 A0A194Q6J4 A0A212FMZ4 A0A2P8Z331 S4PZ13 A0A452FWM8 W5Q4A1 A0A1U7TG25 A0A3Q2HUH5 A0A1S3F2L4 A0A1S3F024 G1Q4G1 A0A384DMA8 A0A3Q7VTS7 A0A3Q7QMU4 A0A3Q7S2Y6 A0A3Q7V2D3 A0A384DM74 A0A3Q7UCR6 A0A384DLL3 A0A3Q7QQ14 A0A2Y9IHL2 A0A3Q7W6U9 A0A3Q7S2Y1 A0A2Y9IGM5 A0A2Y9INX6 A0A2Y9GUB2 A0A2Y9IHL7 A0A3Q7QPJ6 A0A2T3ED75 L8HXH5 A0A452FXE2 M3YLC1 A0A452FWN8 A0A2U3YQS3 A0A3Q1M8C2 A0A3P4M993 A0A1E4Y0N6 A0A0N1I3L0 A0A3Q1BVP7 A0A067QYY2 A0A3Q2Q7P6 A0A146ZRX7 A0A2U3W770 A0A146MT68 A0A146MRF5 A0A146MRI0 A0A248W3R1 A0A3M6U6Z3 M3WX42 A0A1W4YYD7 A0A2J7PTW5 A0A2J7PTW2 A0A2Y9NYC1 A0A3P8XS56 A0A2Y9P1S9 A0A2B4SDX5 A0A340YEQ1 A0A341B8R2 A0A452C6Z3 A0A2J7PTW3 A0A3Q2WUM8 I3JGT7 A0A2J7PTW6 A0A1C3V031 A0A210Q0I0 A0A401SVU4 A0A1H9V393 A0A1C3UG58 A0A3B4ZN00 B3S131 A0A0A8GDC9 A0A2A6JHF4 J0W035 A0A3B4ZL86 A0A369SLN6 A0A3Q3F0M6 A0A369C1E1 C5LW11 A0A0M3GLY8 A0A192T5J0 G3NN69 A0A444HVZ8 A0A1B9SDI1 A0A3Q3W811 A0A372K6B1 A0A3S3WY66 A0A329YPC5 A7SUL1 A0A2U0WLP7
PDB
6EAC
E-value=0.0303087,
Score=80
Ontologies
PANTHER
Topology
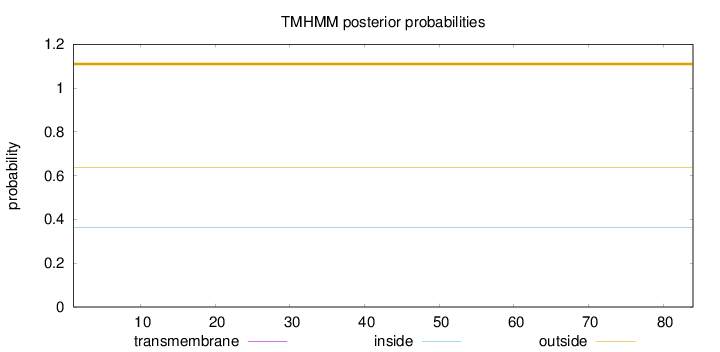
Length:
84
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.36424
outside
1 - 84
Population Genetic Test Statistics
Pi
89.894102
Theta
134.13718
Tajima's D
-0.765584
CLR
199.341593
CSRT
0.179941002949853
Interpretation
Uncertain
