Gene
KWMTBOMO09447
Pre Gene Modal
BGIBMGA012785
Annotation
PREDICTED:_UPF0061_protein_PFL_0486-like_[Papilio_xuthus]
Location in the cell
Extracellular Reliability : 1.329 Mitochondrial Reliability : 1.351
Sequence
CDS
ATGAACGTTTTGATTCAGGCTACTCCGATTCCACTGGAGAATAACGTCAGACTCGTGTGTGTCTCTGTAGATGCACTCGCTAACATTTTAGACATGGATCCGGATGTATCGAATCGAGAGGAATTCGTGGAGTTTGTATCCGGCAGGAAACTTCCTTACGGCGCTTTGCCTATTGCACATCGTTATGGAGGTCATCAATATGGTCTATGGGCTGGCCAACTCGGAGACGGGCGGGCTCATATCATCGGAGAATATGTTAATAGTCGCAATGAGAGATGGCAACTGCAGCTGAAAGGGTCCGGTCCGACGCCGTATTCCCGGATGCGCGACGGTCGCTTGGTGTTACGCGCGGCCATCAGGGAAATGGTGGTCAGTGAAGCCTGCCACTATCTGGGCGTGCCCACAACACGAGCGGCCGCTGTCATCGGTAAGTTGGTTGGATGA
Protein
MNVLIQATPIPLENNVRLVCVSVDALANILDMDPDVSNREEFVEFVSGRKLPYGALPIAHRYGGHQYGLWAGQLGDGRAHIIGEYVNSRNERWQLQLKGSGPTPYSRMRDGRLVLRAAIREMVVSEACHYLGVPTTRAAAVIGKLVG
Summary
Uniprot
H9JTC3
A0A2H1V3U7
A0A194Q0B0
A0A2A4K3S9
A0A194R1J0
A0A3S2LZX5
+ More
A0A212FN18 A0A212FMZ4 A0A3S2NCZ1 A0A2A4K2D8 A0A194Q6J4 A0A067QYY2 U3JXL5 A0A3L8SY93 A0A2U3YQS1 A0A2P8Z331 A0A2I0MR40 V9L1A7 A0A0P7VPH6 A0A146VK85 A0A3Q7S2Y6 A0A452FWN8 W5Q4A1 A0A3Q7QQ14 A0A452FXE2 A0A452FWL2 A0A2U3W770 A0A3Q7S2Z7 A0A3Q7S2Y1 A0A3Q1M8C2 A0A218ULQ4 A0A3Q2HUH5 A0A2D0S4E6 A0A2D4LU69 A0A2Y9INX6 A0A3P4M993 A0A210Q0I0 A0A2Y9IGM5 V8NL78 A0A2H6NIN5 A0A2Y9IHL2 A0A146VKP2 V9KKW6 A0A2H6NFN6 A0A3Q0GQ56 A0A3B4DDI0 A0A3Q0GUX1 A0A1V4KCU1 A0A1V4KBC4 A0A060WQC9 A0A2H6NFM0 A0A2D4LU65 G3WEG1 A0A2D4LU87 G1Q4G1 A0A2D0S2V9 T1HB56 A0A146ZRX7 A0A3Q0GUQ4 L8HXH5 M3WX42 A0A1S3F2L4 A0A3B4ZL28 A0A3B4ZN00 A0A2D4LU43 A0A224XNJ7 A0A3B4ZL86 A0A384DMA8 A0A3Q7VTS7 K7GEL1 A0A3Q7UCR6 A0A384DLL3 A0A3Q2D655 A0A3Q2WUM8 A0A1W4YYD7 A0A3B4F329 A0A3Q2VN24 A0A3P9CT62 A0A3P8P5D2 A0A2Y9EUL2 A0A401SVU4 A0A2Y9EVX9 A0A2R7VSR1 A0A452C6W7 A0A151PCT7 A0A452C6Z3 A0A1S3Q9M3 A0A146MRF5 A0A1S3Q9T8 A0A3Q0GQ92 A0A3P8WZM1 A0A452IJ01 A0A146MT68 E6ZJB8 V3ZJZ7 A0A3Q4GK87 A0A146MRI0 A0A452IJ62 A0A2H1V2H0 A0A1L8FVG7
A0A212FN18 A0A212FMZ4 A0A3S2NCZ1 A0A2A4K2D8 A0A194Q6J4 A0A067QYY2 U3JXL5 A0A3L8SY93 A0A2U3YQS1 A0A2P8Z331 A0A2I0MR40 V9L1A7 A0A0P7VPH6 A0A146VK85 A0A3Q7S2Y6 A0A452FWN8 W5Q4A1 A0A3Q7QQ14 A0A452FXE2 A0A452FWL2 A0A2U3W770 A0A3Q7S2Z7 A0A3Q7S2Y1 A0A3Q1M8C2 A0A218ULQ4 A0A3Q2HUH5 A0A2D0S4E6 A0A2D4LU69 A0A2Y9INX6 A0A3P4M993 A0A210Q0I0 A0A2Y9IGM5 V8NL78 A0A2H6NIN5 A0A2Y9IHL2 A0A146VKP2 V9KKW6 A0A2H6NFN6 A0A3Q0GQ56 A0A3B4DDI0 A0A3Q0GUX1 A0A1V4KCU1 A0A1V4KBC4 A0A060WQC9 A0A2H6NFM0 A0A2D4LU65 G3WEG1 A0A2D4LU87 G1Q4G1 A0A2D0S2V9 T1HB56 A0A146ZRX7 A0A3Q0GUQ4 L8HXH5 M3WX42 A0A1S3F2L4 A0A3B4ZL28 A0A3B4ZN00 A0A2D4LU43 A0A224XNJ7 A0A3B4ZL86 A0A384DMA8 A0A3Q7VTS7 K7GEL1 A0A3Q7UCR6 A0A384DLL3 A0A3Q2D655 A0A3Q2WUM8 A0A1W4YYD7 A0A3B4F329 A0A3Q2VN24 A0A3P9CT62 A0A3P8P5D2 A0A2Y9EUL2 A0A401SVU4 A0A2Y9EVX9 A0A2R7VSR1 A0A452C6W7 A0A151PCT7 A0A452C6Z3 A0A1S3Q9M3 A0A146MRF5 A0A1S3Q9T8 A0A3Q0GQ92 A0A3P8WZM1 A0A452IJ01 A0A146MT68 E6ZJB8 V3ZJZ7 A0A3Q4GK87 A0A146MRI0 A0A452IJ62 A0A2H1V2H0 A0A1L8FVG7
Pubmed
EMBL
BABH01004144
ODYU01000368
SOQ35032.1
KQ459582
KPI99017.1
NWSH01000207
+ More
PCG78423.1 KQ460878 KPJ11663.1 RSAL01000093 RVE47912.1 AGBW02007651 OWR55099.1 OWR55093.1 RVE47911.1 PCG78425.1 KPI99020.1 KK853093 KDR11508.1 AGTO01000113 AGTO01020352 QUSF01000003 RLW11451.1 PYGN01000219 PSN50911.1 AKCR02000004 PKK32153.1 JW872523 AFP05041.1 JARO02001490 KPP75339.1 GCES01068489 JAR17834.1 LWLT01000014 AMGL01024142 MUZQ01000228 OWK54699.1 IACM01047456 LAB24622.1 CYRY02008221 VCW76992.1 NEDP02005302 OWF42250.1 AZIM01003046 ETE62820.1 IACI01091811 LAA31483.1 GCES01068488 JAR17835.1 JW866188 AFO98705.1 IACI01091812 LAA31486.1 LSYS01003958 OPJ81707.1 OPJ81708.1 FR904577 CDQ67244.1 IACI01091810 LAA31481.1 IACM01047459 IACM01047465 LAB24647.1 AEFK01200057 AEFK01200058 AEFK01200059 AEFK01200060 AEFK01200061 AEFK01200062 IACM01047460 IACM01047461 IACM01047463 IACM01047467 LAB24642.1 AAPE02040420 AAPE02040421 AAPE02040422 AAPE02040423 ACPB03016348 GCES01017254 JAR69069.1 JH882683 ELR48546.1 AANG04002256 IACM01047458 LAB24627.1 GFTR01006867 JAW09559.1 AGCU01034247 BEZZ01000605 GCC34501.1 KK854003 PTY08995.1 AKHW03000487 KYO46818.1 GCES01164041 JAQ22281.1 GCES01164040 JAQ22282.1 FQ310508 CBN82152.1 KB202237 ESO91618.1 GCES01164039 JAQ22283.1 SOQ35031.1 CM004476 OCT75579.1
PCG78423.1 KQ460878 KPJ11663.1 RSAL01000093 RVE47912.1 AGBW02007651 OWR55099.1 OWR55093.1 RVE47911.1 PCG78425.1 KPI99020.1 KK853093 KDR11508.1 AGTO01000113 AGTO01020352 QUSF01000003 RLW11451.1 PYGN01000219 PSN50911.1 AKCR02000004 PKK32153.1 JW872523 AFP05041.1 JARO02001490 KPP75339.1 GCES01068489 JAR17834.1 LWLT01000014 AMGL01024142 MUZQ01000228 OWK54699.1 IACM01047456 LAB24622.1 CYRY02008221 VCW76992.1 NEDP02005302 OWF42250.1 AZIM01003046 ETE62820.1 IACI01091811 LAA31483.1 GCES01068488 JAR17835.1 JW866188 AFO98705.1 IACI01091812 LAA31486.1 LSYS01003958 OPJ81707.1 OPJ81708.1 FR904577 CDQ67244.1 IACI01091810 LAA31481.1 IACM01047459 IACM01047465 LAB24647.1 AEFK01200057 AEFK01200058 AEFK01200059 AEFK01200060 AEFK01200061 AEFK01200062 IACM01047460 IACM01047461 IACM01047463 IACM01047467 LAB24642.1 AAPE02040420 AAPE02040421 AAPE02040422 AAPE02040423 ACPB03016348 GCES01017254 JAR69069.1 JH882683 ELR48546.1 AANG04002256 IACM01047458 LAB24627.1 GFTR01006867 JAW09559.1 AGCU01034247 BEZZ01000605 GCC34501.1 KK854003 PTY08995.1 AKHW03000487 KYO46818.1 GCES01164041 JAQ22281.1 GCES01164040 JAQ22282.1 FQ310508 CBN82152.1 KB202237 ESO91618.1 GCES01164039 JAQ22283.1 SOQ35031.1 CM004476 OCT75579.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000053240
UP000283053
UP000007151
+ More
UP000027135 UP000016665 UP000276834 UP000245341 UP000245037 UP000053872 UP000034805 UP000286641 UP000291000 UP000002356 UP000245340 UP000009136 UP000197619 UP000002281 UP000221080 UP000248482 UP000242188 UP000189705 UP000261440 UP000190648 UP000193380 UP000007648 UP000001074 UP000015103 UP000011712 UP000081671 UP000261400 UP000261680 UP000286642 UP000007267 UP000291021 UP000265020 UP000264840 UP000192224 UP000261460 UP000265160 UP000265100 UP000248484 UP000287033 UP000261681 UP000050525 UP000087266 UP000265120 UP000291020 UP000030746 UP000261580 UP000186698
UP000027135 UP000016665 UP000276834 UP000245341 UP000245037 UP000053872 UP000034805 UP000286641 UP000291000 UP000002356 UP000245340 UP000009136 UP000197619 UP000002281 UP000221080 UP000248482 UP000242188 UP000189705 UP000261440 UP000190648 UP000193380 UP000007648 UP000001074 UP000015103 UP000011712 UP000081671 UP000261400 UP000261680 UP000286642 UP000007267 UP000291021 UP000265020 UP000264840 UP000192224 UP000261460 UP000265160 UP000265100 UP000248484 UP000287033 UP000261681 UP000050525 UP000087266 UP000265120 UP000291020 UP000030746 UP000261580 UP000186698
PRIDE
Pfam
PF02696 UPF0061
ProteinModelPortal
H9JTC3
A0A2H1V3U7
A0A194Q0B0
A0A2A4K3S9
A0A194R1J0
A0A3S2LZX5
+ More
A0A212FN18 A0A212FMZ4 A0A3S2NCZ1 A0A2A4K2D8 A0A194Q6J4 A0A067QYY2 U3JXL5 A0A3L8SY93 A0A2U3YQS1 A0A2P8Z331 A0A2I0MR40 V9L1A7 A0A0P7VPH6 A0A146VK85 A0A3Q7S2Y6 A0A452FWN8 W5Q4A1 A0A3Q7QQ14 A0A452FXE2 A0A452FWL2 A0A2U3W770 A0A3Q7S2Z7 A0A3Q7S2Y1 A0A3Q1M8C2 A0A218ULQ4 A0A3Q2HUH5 A0A2D0S4E6 A0A2D4LU69 A0A2Y9INX6 A0A3P4M993 A0A210Q0I0 A0A2Y9IGM5 V8NL78 A0A2H6NIN5 A0A2Y9IHL2 A0A146VKP2 V9KKW6 A0A2H6NFN6 A0A3Q0GQ56 A0A3B4DDI0 A0A3Q0GUX1 A0A1V4KCU1 A0A1V4KBC4 A0A060WQC9 A0A2H6NFM0 A0A2D4LU65 G3WEG1 A0A2D4LU87 G1Q4G1 A0A2D0S2V9 T1HB56 A0A146ZRX7 A0A3Q0GUQ4 L8HXH5 M3WX42 A0A1S3F2L4 A0A3B4ZL28 A0A3B4ZN00 A0A2D4LU43 A0A224XNJ7 A0A3B4ZL86 A0A384DMA8 A0A3Q7VTS7 K7GEL1 A0A3Q7UCR6 A0A384DLL3 A0A3Q2D655 A0A3Q2WUM8 A0A1W4YYD7 A0A3B4F329 A0A3Q2VN24 A0A3P9CT62 A0A3P8P5D2 A0A2Y9EUL2 A0A401SVU4 A0A2Y9EVX9 A0A2R7VSR1 A0A452C6W7 A0A151PCT7 A0A452C6Z3 A0A1S3Q9M3 A0A146MRF5 A0A1S3Q9T8 A0A3Q0GQ92 A0A3P8WZM1 A0A452IJ01 A0A146MT68 E6ZJB8 V3ZJZ7 A0A3Q4GK87 A0A146MRI0 A0A452IJ62 A0A2H1V2H0 A0A1L8FVG7
A0A212FN18 A0A212FMZ4 A0A3S2NCZ1 A0A2A4K2D8 A0A194Q6J4 A0A067QYY2 U3JXL5 A0A3L8SY93 A0A2U3YQS1 A0A2P8Z331 A0A2I0MR40 V9L1A7 A0A0P7VPH6 A0A146VK85 A0A3Q7S2Y6 A0A452FWN8 W5Q4A1 A0A3Q7QQ14 A0A452FXE2 A0A452FWL2 A0A2U3W770 A0A3Q7S2Z7 A0A3Q7S2Y1 A0A3Q1M8C2 A0A218ULQ4 A0A3Q2HUH5 A0A2D0S4E6 A0A2D4LU69 A0A2Y9INX6 A0A3P4M993 A0A210Q0I0 A0A2Y9IGM5 V8NL78 A0A2H6NIN5 A0A2Y9IHL2 A0A146VKP2 V9KKW6 A0A2H6NFN6 A0A3Q0GQ56 A0A3B4DDI0 A0A3Q0GUX1 A0A1V4KCU1 A0A1V4KBC4 A0A060WQC9 A0A2H6NFM0 A0A2D4LU65 G3WEG1 A0A2D4LU87 G1Q4G1 A0A2D0S2V9 T1HB56 A0A146ZRX7 A0A3Q0GUQ4 L8HXH5 M3WX42 A0A1S3F2L4 A0A3B4ZL28 A0A3B4ZN00 A0A2D4LU43 A0A224XNJ7 A0A3B4ZL86 A0A384DMA8 A0A3Q7VTS7 K7GEL1 A0A3Q7UCR6 A0A384DLL3 A0A3Q2D655 A0A3Q2WUM8 A0A1W4YYD7 A0A3B4F329 A0A3Q2VN24 A0A3P9CT62 A0A3P8P5D2 A0A2Y9EUL2 A0A401SVU4 A0A2Y9EVX9 A0A2R7VSR1 A0A452C6W7 A0A151PCT7 A0A452C6Z3 A0A1S3Q9M3 A0A146MRF5 A0A1S3Q9T8 A0A3Q0GQ92 A0A3P8WZM1 A0A452IJ01 A0A146MT68 E6ZJB8 V3ZJZ7 A0A3Q4GK87 A0A146MRI0 A0A452IJ62 A0A2H1V2H0 A0A1L8FVG7
PDB
6EAC
E-value=6.27631e-24,
Score=268
Ontologies
GO
Topology
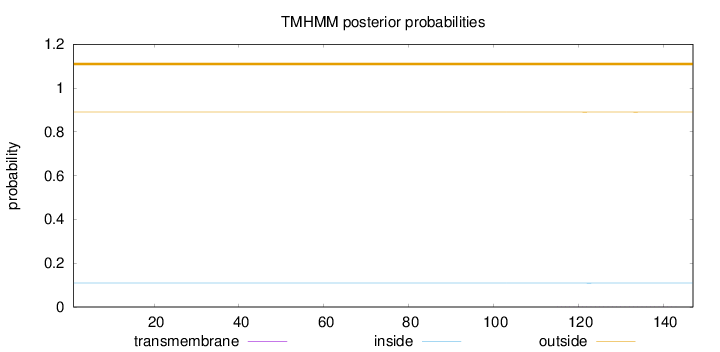
Length:
147
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0154
Exp number, first 60 AAs:
0.00096
Total prob of N-in:
0.10968
outside
1 - 147
Population Genetic Test Statistics
Pi
250.688569
Theta
178.727919
Tajima's D
2.060114
CLR
0.018603
CSRT
0.889705514724264
Interpretation
Uncertain
