Gene
KWMTBOMO09446
Annotation
reverse_transcriptase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.912
Sequence
CDS
ATGTTGCGTTTTGCAGGATCGTTTTGTTATTATTACAGTAGATTGGGGGGAATAGAAACTAGAGTTAGATTAGAGACAAATCTCCCTGGGGGACCGGCTTTCCCCCTGGGGGACCGGCTCCTTGAGCTATATAATGGCTGCTTGGAGTCGGGACGGTTTCCGTCGCTCTGGCGGACGGGTAGACTTGTGTTGTTGAGGAAGGAGGGGCGCCCGGTGGATACAGCCGCCGGGTATCGTCCTATCGTGCTGCTGGACGAGGCGGGAAAATTGCTGGAACGTATTCTGGCTGCCCGCATCGTTCGGCACCTGGTCGGGGTGGGGCCTGACCTGTCGGCGGAGCAGTACGGCTTCCGGGAGGGCCGTTCGACCGTGGATGCAATTCTTCGCGTGCGGTCCCTCTCGGACGAGGCCGTTTCTCGGGGTGGGGTGGCGCTGGCGGTGTCTCTTGACATCGCCAACGCATTTAACACTCTGCCCTGGACCGTGATAGGGGGGGCACTGGAGAGGCATGGAGTGCCCCTCTACCTCCGCCGGCTGGTTGGGTCCTATTTGGGGGCCAGGGGTCGGGGTGCAGTTGAAGTACCTCGGCCTCGTGTTGGACAGCCGGTGGGCCTTTCGTGCTCACTTTGCGGAGCTGGTCCCCGATTGATGGGGACGGCCGGTTCTTTGAGCCGGCTGCTCCCGAATATTGGGGGACCGGATCAGGTTGTGCGCCGTCTCTACGCGGGGGTGGTGCGATCGATGGCCCTGTACGGTGCACCTGTGTGGGCTGAGCCGCGCAACCGGGCCACTATGGCTCGGTTCTTGCGCCGGCCGCAGCGCACCGTTGCCATCAGGGTCATCCGCGGATATCGCACCGTCTCCTTCGAGGCGGCGTGTGTTTTGGCGGGGACGCCGCCATGGGAGCTGGAGGCGGAGTCGCTCGCTGCCGACTATCGGTGGCGCAGCGAGCTTCGTGCTCGGGGCGTGGCGCGTGTCCCCGAGAGTGAGCTGCGGGCGCGGAAGGCCCATTCTCGGCGGTCCGTGCTCGAGTCGTGGTCGAGGCGATTAGCCAACCCCACGTGGGGGCTACGGACCGTCGAGGCGGTTTACCCGGTCTTTGATGACTGGGTGAATCGTGGCGAGGGACGTCTCACCTTTCGTCTGGTGCAGGTGCTGACCGGGCACGGATGCTTCGGGAAGTACCTGCGCCGGATAGGGGCTGAGCCGACGACGAGGTGTCACCATTGTGGACACGACCTGGACACGGCGGAGCATACGCTCGCTGTCTGCCCCGCTTGGGAGGTGCAGCGCCGTGTCCTGGTCGCAAAGATAGGACCTGACTTGTCGCTGCCTGGCGTCGTGGCGTCGATGCTTGGCAGCGATGAGTCATGGAAGGCTATGCTCGACTTCTGCGAGTGCACCATCTCGCAGAAGGAGGCGGCGGGGCGAGTGAGGGAAAGCTCTCCTCACTACGCAGAAACCCGCCGCCGCCGAGCAGGGGGTCGGGACCGGGGTCGTATCCGTGACCTGGCCCCCTAA
Protein
MLRFAGSFCYYYSRLGGIETRVRLETNLPGGPAFPLGDRLLELYNGCLESGRFPSLWRTGRLVLLRKEGRPVDTAAGYRPIVLLDEAGKLLERILAARIVRHLVGVGPDLSAEQYGFREGRSTVDAILRVRSLSDEAVSRGGVALAVSLDIANAFNTLPWTVIGGALERHGVPLYLRRLVGSYLGARGRGAVEVPRPRVGQPVGLSCSLCGAGPRLMGTAGSLSRLLPNIGGPDQVVRRLYAGVVRSMALYGAPVWAEPRNRATMARFLRRPQRTVAIRVIRGYRTVSFEAACVLAGTPPWELEAESLAADYRWRSELRARGVARVPESELRARKAHSRRSVLESWSRRLANPTWGLRTVEAVYPVFDDWVNRGEGRLTFRLVQVLTGHGCFGKYLRRIGAEPTTRCHHCGHDLDTAEHTLAVCPAWEVQRRVLVAKIGPDLSLPGVVASMLGSDESWKAMLDFCECTISQKEAAGRVRESSPHYAETRRRRAGGRDRGRIRDLAP
Summary
Uniprot
Proteomes
Interpro
Gene 3D
ProteinModelPortal
PDB
6AR3
E-value=0.0355702,
Score=88
Ontologies
GO
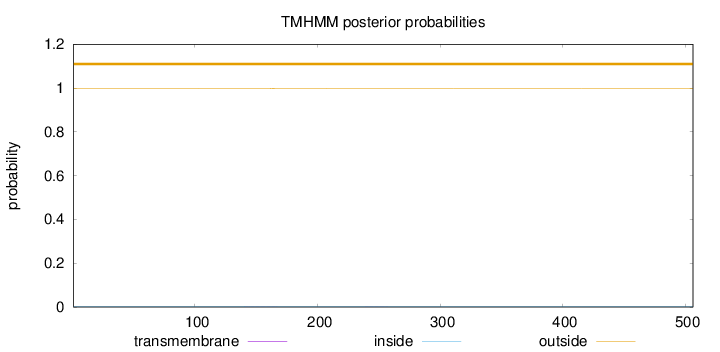
Topology
Length:
506
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02805
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.00184
outside
1 - 506
Population Genetic Test Statistics
Pi
148.8845
Theta
4.866195
Tajima's D
1.53558
CLR
257.347421
CSRT
0.814259287035648
Interpretation
Uncertain
