Gene
KWMTBOMO09445
Pre Gene Modal
BGIBMGA013075
Annotation
PREDICTED:_mevalonate_kinase_isoform_X1_[Bombyx_mori]
Full name
Mevalonate kinase
+ More
Alpha-1,3-glucosyltransferase
Alpha-1,3-glucosyltransferase
Location in the cell
Mitochondrial Reliability : 1.901
Sequence
CDS
ATGTTGACGGCCGTCGATAATTTGTTTACGGTGAGCGTTTCCGCGCCGGGGAAAGTCATCTTGCATGGAGAGCATTCGGTCGTGTACGGAAAAACTGCGATCGCTGTCAGCCTCGGCCTAAGGAGCTCCATAGTGATAAAGGAAGTAAACACTCCACACGAACCAGCAGTCCACATACATTTGCCGTGCGTGGATCTACAGGAGACCATTCCATTGGAGCCGACAGTCAAAAGTCTGTTCCATCCTAAACTGGCACCAGGGATAACCGGCAAATTCTCTTGGAGATTACCCCACAAAATCGACCATGACTATCACTTGCGAAGGGTGGAGGAATACCTGCATTTGATCAAGCCTAACTTTGACTCGTTGCCCAACAATCAAAAGAATTCCTTGCGTAGTTTCCTATACGTGTTTTCCGGGATTTTCGGTAGCACATATCTACCGGTAAAGTCTATGGATATTTCGATGGGATCTGAGCTCACCATTGGCGCGGGTACTGGCAGTTCGGCGTCTTTTGCCGTCTGTCTAGCAGGAGCATTGATACAGCTGTTGAAATTGAAGAGTTCTTCGGGGAATTTCGATGCATATTACGATCAGAGCAGCCAAGATTTTACGGATACCGAAAAGGAGATTATATCGGAATGGGCCTTCAATTGTGAGAAGATAATGCACGGAACACCTTCAGGTATTGACAACGCGACCTGCACATTCGGGAACTTGGTATCGTTCAAGAAAGGCGCTAAACCACGTCATCTCGACATAAGGATGGAGCTGCGCGTGTTGCTGGTCGATTCGCGGGTGTCCAGAGAAACGAGGACCCTGGTCGTTCGTGTGGCGGCCCTAAGGCAAAGGAACACGGCCGCCGTGGACCACATCATGGAGGCCTGTGAACACGTTGCGCACACTGCCACTCAGGTTTTAGAGAAACTTTCCAGCGGCAAGTGTGACCCCGATACGGAGGCGGACTATCAACATTTATCAGAGCTTTGGGACATGAACCACTGTCTGCTGTCGGCTCTGGGCGTGTCTCATCCCTCTCTCGAGGTGATCCGAGCGGCGGCCAAGAGCAAGGGGCTCGCCTGCAAGCTCACCGGAGCCGGAGGTGGAGGATACGCCATGATCTTAATTCCACCATCGACTCCTCGTTCGACGATAGACTCGCTGTCCGGCCAGCTGCTGCAGAACGGTTTCCGGGTCACGGAGACGAGGCTCGGTGGACCGGGCGTCTCCATAGAGATGTGA
Protein
MLTAVDNLFTVSVSAPGKVILHGEHSVVYGKTAIAVSLGLRSSIVIKEVNTPHEPAVHIHLPCVDLQETIPLEPTVKSLFHPKLAPGITGKFSWRLPHKIDHDYHLRRVEEYLHLIKPNFDSLPNNQKNSLRSFLYVFSGIFGSTYLPVKSMDISMGSELTIGAGTGSSASFAVCLAGALIQLLKLKSSSGNFDAYYDQSSQDFTDTEKEIISEWAFNCEKIMHGTPSGIDNATCTFGNLVSFKKGAKPRHLDIRMELRVLLVDSRVSRETRTLVVRVAALRQRNTAAVDHIMEACEHVAHTATQVLEKLSSGKCDPDTEADYQHLSELWDMNHCLLSALGVSHPSLEVIRAAAKSKGLACKLTGAGGGGYAMILIPPSTPRSTIDSLSGQLLQNGFRVTETRLGGPGVSIEM
Summary
Description
Catalyzes the phosphorylation of mevalonate to mevalonate 5-phosphate, a key step in isoprenoid and cholesterol biosynthesis.
Catalytic Activity
(R)-mevalonate + ATP = (R)-5-phosphomevalonate + ADP + H(+)
Similarity
Belongs to the GHMP kinase family. Mevalonate kinase subfamily.
Belongs to the ALG6/ALG8 glucosyltransferase family.
Belongs to the ALG6/ALG8 glucosyltransferase family.
Feature
chain Alpha-1,3-glucosyltransferase
Uniprot
A5A7A0
A0A2H1VIA5
A0A2A4K2D1
A0A2H4UZK0
H9JU63
A0A437BC38
+ More
A0A212FMY0 I4DKG6 A0A194Q6I9 A0A194R717 A0A0H4IUW0 A0A2P8Z343 A0A067RSI1 A0A0L7REB1 A0A310SIZ3 A0A2J7PTV6 K7IN15 A0A088A8H7 A0A1B0CYD0 A0A2A3EGN3 I1VX02 A0A0G3F5W2 A0A1L8DZ74 A0A1W4XCS0 I1VJ72 A0A232F1I2 A0A1B6IB16 A0A1Y1MYH1 D2A4R0 A0A1B6C337 E2AQ72 A0A0J7L7Q1 A0A195FPX4 U4UN04 A0A195CCX2 A0A154PLJ5 Q176C1 A0A077WT23 U5EVK9 A0A1A7WQ55 A0A0A9XFL2 A0A0P4VUR7 A0A1S3HMS2 N6UI58 A0A3L8D9P7 A0A1J7JK90 E2BLU0 A0A0V0G886 A0A023ESL4 A0A158NEU5 A0A1Y2U394 A0A1Y2USH0 A0A151WSI6 B4MJK1 C1BQV3 A0A162NK42 T1PGP7 V4A0T9 A0A168QSE1 A0A2T7PHZ0 A0A1I8N5P9 F4WJE7 T1IDA7 A0A1A8FM48 E9IEK9 A0A1B0AAW7 A0A194X6X2 A0A165I9M8 A0A1Y2W8M0 A0A397TW19 A0A1A9V2U3 D3TMC0 B2B6Y4 A0A146P6P7 A0A3Q4BQL6 A0A2N1MUP2 A0A3Q3JIA9 F7VU92 A0A0S7I2W7 A0A3P8XDF1 A0A367JTM1 A0A0A1N2Y4 A0A1B5L0M7 A0A1A8BX20 A7SJK4 A0A096M390 A0A3B3V7P1 A0A015L7P9 G2Q8J6 A0A3B5AZV0 A0A146P4D7 B3NRX6 A0A146NPZ2 A0A3B5MEV8 A0A3Q2CCL6 A0A2H5TIE6 A0A0F4ZDB1 A0A0F8AY33 A0A3B5KC85
A0A212FMY0 I4DKG6 A0A194Q6I9 A0A194R717 A0A0H4IUW0 A0A2P8Z343 A0A067RSI1 A0A0L7REB1 A0A310SIZ3 A0A2J7PTV6 K7IN15 A0A088A8H7 A0A1B0CYD0 A0A2A3EGN3 I1VX02 A0A0G3F5W2 A0A1L8DZ74 A0A1W4XCS0 I1VJ72 A0A232F1I2 A0A1B6IB16 A0A1Y1MYH1 D2A4R0 A0A1B6C337 E2AQ72 A0A0J7L7Q1 A0A195FPX4 U4UN04 A0A195CCX2 A0A154PLJ5 Q176C1 A0A077WT23 U5EVK9 A0A1A7WQ55 A0A0A9XFL2 A0A0P4VUR7 A0A1S3HMS2 N6UI58 A0A3L8D9P7 A0A1J7JK90 E2BLU0 A0A0V0G886 A0A023ESL4 A0A158NEU5 A0A1Y2U394 A0A1Y2USH0 A0A151WSI6 B4MJK1 C1BQV3 A0A162NK42 T1PGP7 V4A0T9 A0A168QSE1 A0A2T7PHZ0 A0A1I8N5P9 F4WJE7 T1IDA7 A0A1A8FM48 E9IEK9 A0A1B0AAW7 A0A194X6X2 A0A165I9M8 A0A1Y2W8M0 A0A397TW19 A0A1A9V2U3 D3TMC0 B2B6Y4 A0A146P6P7 A0A3Q4BQL6 A0A2N1MUP2 A0A3Q3JIA9 F7VU92 A0A0S7I2W7 A0A3P8XDF1 A0A367JTM1 A0A0A1N2Y4 A0A1B5L0M7 A0A1A8BX20 A7SJK4 A0A096M390 A0A3B3V7P1 A0A015L7P9 G2Q8J6 A0A3B5AZV0 A0A146P4D7 B3NRX6 A0A146NPZ2 A0A3B5MEV8 A0A3Q2CCL6 A0A2H5TIE6 A0A0F4ZDB1 A0A0F8AY33 A0A3B5KC85
EC Number
2.7.1.36
2.4.1.-
2.4.1.-
Pubmed
17628279
29185447
19121390
22118469
22651552
26354079
+ More
26899871 29403074 24845553 20075255 22516182 28648823 28004739 18362917 19820115 20798317 23537049 17510324 25401762 26823975 27129103 30249741 24945155 21347285 28078400 17994087 23254933 25315136 21719571 21282665 26693682 20353571 18460219 24558260 20386741 25069045 29674435 27956601 17615350 21964414 25835551 21551351
26899871 29403074 24845553 20075255 22516182 28648823 28004739 18362917 19820115 20798317 23537049 17510324 25401762 26823975 27129103 30249741 24945155 21347285 28078400 17994087 23254933 25315136 21719571 21282665 26693682 20353571 18460219 24558260 20386741 25069045 29674435 27956601 17615350 21964414 25835551 21551351
EMBL
AB274991
BAF62109.1
ODYU01002714
SOQ40569.1
NWSH01000207
PCG78421.1
+ More
KY971768 AUA18035.1 BABH01004151 RSAL01000093 RVE47914.1 AGBW02007651 OWR55101.1 AK401784 BAM18406.1 KQ459582 KPI99015.1 KQ460878 KPJ11661.1 KP689338 AKO63320.1 PYGN01000219 PSN50908.1 KK852445 KDR23770.1 KQ414609 KOC69312.1 KQ767320 OAD53412.1 NEVH01021221 PNF19768.1 AJVK01000004 AJVK01000005 AJVK01000006 KZ288254 PBC30887.1 JQ413984 AFI55100.1 KP420146 AKJ80166.1 GFDF01002340 JAV11744.1 JQ855693 AFI45056.1 NNAY01001310 OXU24402.1 GECU01023571 JAS84135.1 GEZM01023419 JAV88397.1 KQ971344 EFA05256.2 GEDC01029638 JAS07660.1 GL441701 EFN64406.1 LBMM01000361 KMQ99398.1 KQ981335 KYN42645.1 KB632429 ERL95469.1 KQ978023 KYM97933.1 KQ434972 KZC12739.1 CH477390 EAT41980.1 LK023335 CDS10404.1 GANO01001816 JAB58055.1 HADW01006478 SBP07878.1 GBHO01024840 GBRD01003593 GDHC01015377 JAG18764.1 JAG62228.1 JAQ03252.1 GDKW01000132 JAI56463.1 APGK01019085 KB740092 ENN81425.1 QOIP01000011 RLU17056.1 KV875097 OIW30256.1 GL449036 EFN83424.1 GECL01002003 JAP04121.1 GAPW01001827 JAC11771.1 ADTU01013171 KZ111238 OTA67866.1 KZ112560 OTA87273.1 KQ982769 KYQ50869.1 CH963846 EDW72290.1 BT076982 ACO11406.1 KV440988 OAD70414.1 KA647874 AFP62503.1 KB201305 ESO97418.1 LT554468 SAM05448.1 PZQS01000004 PVD33046.1 GL888182 EGI65725.1 ACPB03002946 HAEB01013294 SBQ59821.1 GL762647 EFZ20993.1 KQ947418 KUJ15552.1 KV407456 KZF24589.1 KZ111538 OTB06135.1 QKYT01000007 RIA99064.1 CCAG010016559 EZ422572 ADD18848.1 CU640366 FO904937 CAP73562.1 CDP25965.1 GCES01146804 JAQ39518.1 LLXL01001281 PKK65339.1 CABT02000007 CCC09080.1 GBYX01434783 JAO46546.1 PJQL01000712 RCH93322.1 CDGI01000202 KV921481 CEI93702.1 ORE14256.1 BBTG02000044 GAO16958.1 HADZ01007412 SBP71353.1 DS469678 EDO36115.1 AYCK01007734 AYCK01007735 JEMT01029766 EXX50833.1 CP003003 AEO56245.1 GCES01147491 JAQ38831.1 CH954179 EDV56278.1 GCES01152463 JAQ33859.1 BDIQ01000130 GBC42215.1 LAEV01001554 KKA27858.1 KQ042291 KKF16217.1
KY971768 AUA18035.1 BABH01004151 RSAL01000093 RVE47914.1 AGBW02007651 OWR55101.1 AK401784 BAM18406.1 KQ459582 KPI99015.1 KQ460878 KPJ11661.1 KP689338 AKO63320.1 PYGN01000219 PSN50908.1 KK852445 KDR23770.1 KQ414609 KOC69312.1 KQ767320 OAD53412.1 NEVH01021221 PNF19768.1 AJVK01000004 AJVK01000005 AJVK01000006 KZ288254 PBC30887.1 JQ413984 AFI55100.1 KP420146 AKJ80166.1 GFDF01002340 JAV11744.1 JQ855693 AFI45056.1 NNAY01001310 OXU24402.1 GECU01023571 JAS84135.1 GEZM01023419 JAV88397.1 KQ971344 EFA05256.2 GEDC01029638 JAS07660.1 GL441701 EFN64406.1 LBMM01000361 KMQ99398.1 KQ981335 KYN42645.1 KB632429 ERL95469.1 KQ978023 KYM97933.1 KQ434972 KZC12739.1 CH477390 EAT41980.1 LK023335 CDS10404.1 GANO01001816 JAB58055.1 HADW01006478 SBP07878.1 GBHO01024840 GBRD01003593 GDHC01015377 JAG18764.1 JAG62228.1 JAQ03252.1 GDKW01000132 JAI56463.1 APGK01019085 KB740092 ENN81425.1 QOIP01000011 RLU17056.1 KV875097 OIW30256.1 GL449036 EFN83424.1 GECL01002003 JAP04121.1 GAPW01001827 JAC11771.1 ADTU01013171 KZ111238 OTA67866.1 KZ112560 OTA87273.1 KQ982769 KYQ50869.1 CH963846 EDW72290.1 BT076982 ACO11406.1 KV440988 OAD70414.1 KA647874 AFP62503.1 KB201305 ESO97418.1 LT554468 SAM05448.1 PZQS01000004 PVD33046.1 GL888182 EGI65725.1 ACPB03002946 HAEB01013294 SBQ59821.1 GL762647 EFZ20993.1 KQ947418 KUJ15552.1 KV407456 KZF24589.1 KZ111538 OTB06135.1 QKYT01000007 RIA99064.1 CCAG010016559 EZ422572 ADD18848.1 CU640366 FO904937 CAP73562.1 CDP25965.1 GCES01146804 JAQ39518.1 LLXL01001281 PKK65339.1 CABT02000007 CCC09080.1 GBYX01434783 JAO46546.1 PJQL01000712 RCH93322.1 CDGI01000202 KV921481 CEI93702.1 ORE14256.1 BBTG02000044 GAO16958.1 HADZ01007412 SBP71353.1 DS469678 EDO36115.1 AYCK01007734 AYCK01007735 JEMT01029766 EXX50833.1 CP003003 AEO56245.1 GCES01147491 JAQ38831.1 CH954179 EDV56278.1 GCES01152463 JAQ33859.1 BDIQ01000130 GBC42215.1 LAEV01001554 KKA27858.1 KQ042291 KKF16217.1
Proteomes
UP000218220
UP000005204
UP000283053
UP000007151
UP000053268
UP000053240
+ More
UP000245037 UP000027135 UP000053825 UP000235965 UP000002358 UP000005203 UP000092462 UP000242457 UP000192223 UP000215335 UP000007266 UP000000311 UP000036403 UP000078541 UP000030742 UP000078542 UP000076502 UP000008820 UP000085678 UP000019118 UP000279307 UP000182658 UP000008237 UP000005205 UP000194279 UP000194361 UP000075809 UP000007798 UP000077315 UP000030746 UP000078561 UP000245119 UP000095301 UP000007755 UP000015103 UP000092445 UP000070700 UP000076632 UP000242955 UP000265703 UP000078200 UP000092444 UP000001197 UP000261620 UP000233469 UP000261600 UP000001881 UP000265140 UP000252139 UP000038169 UP000242381 UP000001593 UP000028760 UP000261500 UP000022910 UP000007322 UP000261400 UP000008711 UP000261380 UP000265020 UP000236242 UP000005226
UP000245037 UP000027135 UP000053825 UP000235965 UP000002358 UP000005203 UP000092462 UP000242457 UP000192223 UP000215335 UP000007266 UP000000311 UP000036403 UP000078541 UP000030742 UP000078542 UP000076502 UP000008820 UP000085678 UP000019118 UP000279307 UP000182658 UP000008237 UP000005205 UP000194279 UP000194361 UP000075809 UP000007798 UP000077315 UP000030746 UP000078561 UP000245119 UP000095301 UP000007755 UP000015103 UP000092445 UP000070700 UP000076632 UP000242955 UP000265703 UP000078200 UP000092444 UP000001197 UP000261620 UP000233469 UP000261600 UP000001881 UP000265140 UP000252139 UP000038169 UP000242381 UP000001593 UP000028760 UP000261500 UP000022910 UP000007322 UP000261400 UP000008711 UP000261380 UP000265020 UP000236242 UP000005226
Interpro
IPR020568
Ribosomal_S5_D2-typ_fold
+ More
IPR006205 Mev_gal_kin
IPR013750 GHMP_kinase_C_dom
IPR014721 Ribosomal_S5_D2-typ_fold_subgr
IPR006204 GHMP_kinase_N_dom
IPR036554 GHMP_kinase_C_sf
IPR039488 ALG6
IPR004856 Glyco_trans_ALG6/ALG8
IPR006203 GHMP_knse_ATP-bd_CS
IPR019539 GalKase_gal-bd
IPR007397 F-box-assoc_dom
IPR001202 WW_dom
IPR006205 Mev_gal_kin
IPR013750 GHMP_kinase_C_dom
IPR014721 Ribosomal_S5_D2-typ_fold_subgr
IPR006204 GHMP_kinase_N_dom
IPR036554 GHMP_kinase_C_sf
IPR039488 ALG6
IPR004856 Glyco_trans_ALG6/ALG8
IPR006203 GHMP_knse_ATP-bd_CS
IPR019539 GalKase_gal-bd
IPR007397 F-box-assoc_dom
IPR001202 WW_dom
Gene 3D
CDD
ProteinModelPortal
A5A7A0
A0A2H1VIA5
A0A2A4K2D1
A0A2H4UZK0
H9JU63
A0A437BC38
+ More
A0A212FMY0 I4DKG6 A0A194Q6I9 A0A194R717 A0A0H4IUW0 A0A2P8Z343 A0A067RSI1 A0A0L7REB1 A0A310SIZ3 A0A2J7PTV6 K7IN15 A0A088A8H7 A0A1B0CYD0 A0A2A3EGN3 I1VX02 A0A0G3F5W2 A0A1L8DZ74 A0A1W4XCS0 I1VJ72 A0A232F1I2 A0A1B6IB16 A0A1Y1MYH1 D2A4R0 A0A1B6C337 E2AQ72 A0A0J7L7Q1 A0A195FPX4 U4UN04 A0A195CCX2 A0A154PLJ5 Q176C1 A0A077WT23 U5EVK9 A0A1A7WQ55 A0A0A9XFL2 A0A0P4VUR7 A0A1S3HMS2 N6UI58 A0A3L8D9P7 A0A1J7JK90 E2BLU0 A0A0V0G886 A0A023ESL4 A0A158NEU5 A0A1Y2U394 A0A1Y2USH0 A0A151WSI6 B4MJK1 C1BQV3 A0A162NK42 T1PGP7 V4A0T9 A0A168QSE1 A0A2T7PHZ0 A0A1I8N5P9 F4WJE7 T1IDA7 A0A1A8FM48 E9IEK9 A0A1B0AAW7 A0A194X6X2 A0A165I9M8 A0A1Y2W8M0 A0A397TW19 A0A1A9V2U3 D3TMC0 B2B6Y4 A0A146P6P7 A0A3Q4BQL6 A0A2N1MUP2 A0A3Q3JIA9 F7VU92 A0A0S7I2W7 A0A3P8XDF1 A0A367JTM1 A0A0A1N2Y4 A0A1B5L0M7 A0A1A8BX20 A7SJK4 A0A096M390 A0A3B3V7P1 A0A015L7P9 G2Q8J6 A0A3B5AZV0 A0A146P4D7 B3NRX6 A0A146NPZ2 A0A3B5MEV8 A0A3Q2CCL6 A0A2H5TIE6 A0A0F4ZDB1 A0A0F8AY33 A0A3B5KC85
A0A212FMY0 I4DKG6 A0A194Q6I9 A0A194R717 A0A0H4IUW0 A0A2P8Z343 A0A067RSI1 A0A0L7REB1 A0A310SIZ3 A0A2J7PTV6 K7IN15 A0A088A8H7 A0A1B0CYD0 A0A2A3EGN3 I1VX02 A0A0G3F5W2 A0A1L8DZ74 A0A1W4XCS0 I1VJ72 A0A232F1I2 A0A1B6IB16 A0A1Y1MYH1 D2A4R0 A0A1B6C337 E2AQ72 A0A0J7L7Q1 A0A195FPX4 U4UN04 A0A195CCX2 A0A154PLJ5 Q176C1 A0A077WT23 U5EVK9 A0A1A7WQ55 A0A0A9XFL2 A0A0P4VUR7 A0A1S3HMS2 N6UI58 A0A3L8D9P7 A0A1J7JK90 E2BLU0 A0A0V0G886 A0A023ESL4 A0A158NEU5 A0A1Y2U394 A0A1Y2USH0 A0A151WSI6 B4MJK1 C1BQV3 A0A162NK42 T1PGP7 V4A0T9 A0A168QSE1 A0A2T7PHZ0 A0A1I8N5P9 F4WJE7 T1IDA7 A0A1A8FM48 E9IEK9 A0A1B0AAW7 A0A194X6X2 A0A165I9M8 A0A1Y2W8M0 A0A397TW19 A0A1A9V2U3 D3TMC0 B2B6Y4 A0A146P6P7 A0A3Q4BQL6 A0A2N1MUP2 A0A3Q3JIA9 F7VU92 A0A0S7I2W7 A0A3P8XDF1 A0A367JTM1 A0A0A1N2Y4 A0A1B5L0M7 A0A1A8BX20 A7SJK4 A0A096M390 A0A3B3V7P1 A0A015L7P9 G2Q8J6 A0A3B5AZV0 A0A146P4D7 B3NRX6 A0A146NPZ2 A0A3B5MEV8 A0A3Q2CCL6 A0A2H5TIE6 A0A0F4ZDB1 A0A0F8AY33 A0A3B5KC85
PDB
2R3V
E-value=6.69709e-40,
Score=412
Ontologies
PATHWAY
GO
PANTHER
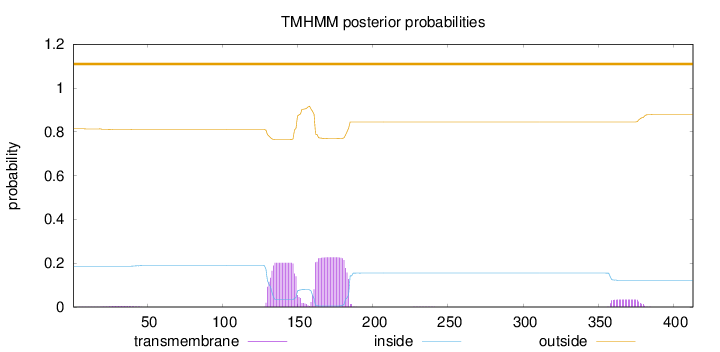
Topology
Subcellular location
Cytoplasm
Endoplasmic reticulum membrane
Endoplasmic reticulum membrane
Length:
413
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
9.74325
Exp number, first 60 AAs:
0.08355
Total prob of N-in:
0.18636
outside
1 - 413
Population Genetic Test Statistics
Pi
21.046323
Theta
21.680505
Tajima's D
0.472764
CLR
1.043936
CSRT
0.508574571271436
Interpretation
Uncertain
