Pre Gene Modal
BGIBMGA012784
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_two_pore_calcium_channel_protein_1-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.578
Sequence
CDS
ATGGGCAGTCGCTCGTCCCTCCACACGGGCCTCCACATGTCCGAAGACGAGCATTGGGAGATGAACTACCACGAGGCAGCTATTTATTTAGAGGAAGGTCAAAACAACGAAAAGTTCGACTCCCATCCATCTAGTCCCGAAGAGTTACCAGCCTACCTCAGGGTACACAACCCCTGGTACCATGGCCTCGATCTACTAGCTTCATTAGTTTTAATATTATTGGCATTCACCGAAGACCCAGCCGTTCCCACATTTGAGCTTCCAGTATGGGCACACGGTACTATAGAATTAATGGCGTTAACTGTGATCGGAATAGAGCTACATCTGAAACTAAAATGGATCGGATGGAGCACCATACTTAAACACAAGAGGACCATGTTAAAGGGTATAACCCTGCTAATAATGGTGCTTGAAGCTGTGGTAGTGCTATGCCGGCAATCCTCACACTTCAGAGTAACGAGGGCGTTGCGTCCTATCTTCTTAGTGGACACGCGGCACTGCGGCGGAGTACGGAGGTTCATTAGACAAATTCTGCAATCGTTACCACCGATTATTGATATGTTAGGTCTATTAATGTTCTTCGTGGCGACTTACTCGCTTTTAGGATTCTATTTATTCTCTGAGCACGTAGACAATGGACACTTCCAGACGCTTAGTGACTCATTCGTTAGTATGTTTGTTTTGCTGACTTCGGCAAACTTCCCAGATGTGATGATGCCATCTTATGCCAAGTCGAAGTGGTACGCACTATTCTTCATCTTGTACATAATCACCGTTCTCTACGTCTTGATGAATTTGATGTTGGCCGTTGTTTACGAGACCTTCACCCGGATCGAACGCGAGAAATGTCGCGCGCTGTTACTGCATCGCCGCGGCGCTACACGTCACGCCTTCCGACTGCTTGTCTCACGACGAGCGCCTCACGCAGTCCGGTTGAGACACTTCGCGGGACTCATGAGACATTACGCGCCGCATTACAATGGTCTAGACGTGTACCTAATGTTCAAGCACCTGAACCAGACCGGCACAGGGGGTCTCTCCCGGGCGGAGTTCGCGAACGTGTACGATGTGTTCGCACTGCGCTGGGCGGCGCAGACCGCGCGCGCTCCGTGGTACGCCGCGTCGCCGCTCGAGCCCATCGCCCGCGCCGCCGCCGCCGCCGTGCACTGGAAACACTTTGAGCATCTTGTCTGTAAGTCCACAGATTACTAA
Protein
MGSRSSLHTGLHMSEDEHWEMNYHEAAIYLEEGQNNEKFDSHPSSPEELPAYLRVHNPWYHGLDLLASLVLILLAFTEDPAVPTFELPVWAHGTIELMALTVIGIELHLKLKWIGWSTILKHKRTMLKGITLLIMVLEAVVVLCRQSSHFRVTRALRPIFLVDTRHCGGVRRFIRQILQSLPPIIDMLGLLMFFVATYSLLGFYLFSEHVDNGHFQTLSDSFVSMFVLLTSANFPDVMMPSYAKSKWYALFFILYIITVLYVLMNLMLAVVYETFTRIEREKCRALLLHRRGATRHAFRLLVSRRAPHAVRLRHFAGLMRHYAPHYNGLDVYLMFKHLNQTGTGGLSRAEFANVYDVFALRWAAQTARAPWYAASPLEPIARAAAAAVHWKHFEHLVCKSTDY
Summary
Uniprot
A0A212FMX6
A0A194R2B4
A0A437BBZ4
A0A2H1VIA3
A0A2A4K419
A0A2A4K2Z5
+ More
A0A2A4K3C1 A0A0L7LMU2 A0A194Q1Z0 A0A2J7PTT9 A0A067QPX2 A0A2P8Z326 A0A2J7PTU8 A0A2J7PTU6 A0A1B6DU32 A0A0C9QCA4 A0A1B6MEL7 D2A6B3 A0A1B6DE94 K7IN16 A0A232F1S9 A0A088A8Q4 A0A2A3EGQ6 A0A1Y1NBW7 A0A1Y1NFP5 A0A310SA12 E0VHQ9 A0A154PN53 E9IEK8 A0A1W4WI41 A0A026WWZ4 E2AQ73 A0A1W4WSY0 A0A158NEU4 A0A195BVH7 E2BLT9 A0A195FRV6 A0A0J7L7X6 U4UR78 A0A151WSM0 F4WJE8 N6TYV1 A0A2S2QQC8 J9K983 A0A0T6BI11 A0A1B6IRK8 A0A0P5LQC5 A0A0P5W0M2 A0A0P6DAA0 A0A0P5K371 A0A0P5A761 A0A0N7ZVX8 A0A0P5TS73 A0A0P4XWY9 A0A0P5BRX1 A0A0N8B6F7 A0A0P5A5V7 A0A0P4ZEA0 A0A0P5WQ39 A0A0N8AQ54 A0A0P5S0P0 A0A0P5M5U4 A0A0P5NLE8 A0A0P5BDU6 A0A0P5LE06 A0A0P5WGR0 A0A0P5V3X6 A0A0P5QI87 A0A0P5DE72 A0A0P6I5P3 A0A0N8CCU7 E9GLT0 A0A0N8DMW7 A0A0P6GTU1 A0A0P5UXL4 A0A3S3Q4H4 A0A0P5LGN1 A0A210Q0H2 D1J6X7 G3T3S0 G3TRD5 B8PXJ8 A0A2G8KRI8 A0A401SP53 A0A3Q7QIU4 A0A2Y9HY96 D2HS01 A0A1S3H6H9 A0A401NPM2 H0XD96 A0A3B4B558 K9IU16 A0A3Q7RXX2 A0A3Q3M710 G1LQ73 W5MPH5 A0A383Z701 A0A383Z7D9 A0A2Y9EMS6
A0A2A4K3C1 A0A0L7LMU2 A0A194Q1Z0 A0A2J7PTT9 A0A067QPX2 A0A2P8Z326 A0A2J7PTU8 A0A2J7PTU6 A0A1B6DU32 A0A0C9QCA4 A0A1B6MEL7 D2A6B3 A0A1B6DE94 K7IN16 A0A232F1S9 A0A088A8Q4 A0A2A3EGQ6 A0A1Y1NBW7 A0A1Y1NFP5 A0A310SA12 E0VHQ9 A0A154PN53 E9IEK8 A0A1W4WI41 A0A026WWZ4 E2AQ73 A0A1W4WSY0 A0A158NEU4 A0A195BVH7 E2BLT9 A0A195FRV6 A0A0J7L7X6 U4UR78 A0A151WSM0 F4WJE8 N6TYV1 A0A2S2QQC8 J9K983 A0A0T6BI11 A0A1B6IRK8 A0A0P5LQC5 A0A0P5W0M2 A0A0P6DAA0 A0A0P5K371 A0A0P5A761 A0A0N7ZVX8 A0A0P5TS73 A0A0P4XWY9 A0A0P5BRX1 A0A0N8B6F7 A0A0P5A5V7 A0A0P4ZEA0 A0A0P5WQ39 A0A0N8AQ54 A0A0P5S0P0 A0A0P5M5U4 A0A0P5NLE8 A0A0P5BDU6 A0A0P5LE06 A0A0P5WGR0 A0A0P5V3X6 A0A0P5QI87 A0A0P5DE72 A0A0P6I5P3 A0A0N8CCU7 E9GLT0 A0A0N8DMW7 A0A0P6GTU1 A0A0P5UXL4 A0A3S3Q4H4 A0A0P5LGN1 A0A210Q0H2 D1J6X7 G3T3S0 G3TRD5 B8PXJ8 A0A2G8KRI8 A0A401SP53 A0A3Q7QIU4 A0A2Y9HY96 D2HS01 A0A1S3H6H9 A0A401NPM2 H0XD96 A0A3B4B558 K9IU16 A0A3Q7RXX2 A0A3Q3M710 G1LQ73 W5MPH5 A0A383Z701 A0A383Z7D9 A0A2Y9EMS6
Pubmed
EMBL
AGBW02007651
OWR55102.1
KQ460878
KPJ11659.1
RSAL01000093
RVE47915.1
+ More
ODYU01002714 SOQ40567.1 NWSH01000207 PCG78420.1 PCG78419.1 PCG78418.1 JTDY01000502 KOB76873.1 KQ459582 KPI99014.1 NEVH01021221 PNF19756.1 KK853093 KDR11510.1 PYGN01000219 PSN50906.1 PNF19757.1 PNF19755.1 GEDC01008120 JAS29178.1 GBYB01000919 JAG70686.1 GEBQ01005597 JAT34380.1 KQ971347 EFA05489.1 GEDC01013338 JAS23960.1 NNAY01001310 OXU24398.1 KZ288254 PBC30888.1 GEZM01007055 JAV95444.1 GEZM01007054 JAV95445.1 KQ767320 OAD53411.1 DS235171 EEB12945.1 KQ434972 KZC12738.1 GL762647 EFZ20995.1 KK107078 QOIP01000011 EZA60358.1 RLU16920.1 GL441701 EFN64407.1 ADTU01013171 KQ976401 KYM92587.1 GL449036 EFN83423.1 KQ981335 KYN42644.1 LBMM01000361 KMQ99399.1 KB632330 ERL92570.1 KQ982769 KYQ50870.1 GL888182 EGI65726.1 APGK01055716 APGK01055717 KB741269 ENN71457.1 GGMS01010776 MBY79979.1 ABLF02033059 ABLF02033063 LJIG01000129 KRT86811.1 GECU01018154 JAS89552.1 GDIQ01166711 JAK85014.1 GDIP01092541 JAM11174.1 GDIQ01081934 JAN12803.1 GDIQ01191592 JAK60133.1 GDIP01203162 JAJ20240.1 GDIP01207313 JAJ16089.1 GDIP01122333 JAL81381.1 GDIP01235565 JAI87836.1 GDIP01181671 JAJ41731.1 GDIQ01253826 GDIQ01252532 GDIQ01251187 GDIQ01208203 GDIQ01206598 GDIQ01205148 GDIP01091575 LRGB01000944 JAK43522.1 JAM12140.1 KZS14480.1 GDIP01216023 JAJ07379.1 GDIP01218171 JAJ05231.1 GDIP01083883 JAM19832.1 GDIQ01253827 GDIQ01252533 GDIQ01251188 GDIQ01248042 GDIQ01208204 GDIQ01206599 GDIQ01205149 GDIQ01203451 JAJ97897.1 GDIQ01192649 GDIQ01094534 JAL57192.1 GDIQ01209199 GDIQ01201697 GDIQ01184354 GDIQ01084837 JAK67371.1 GDIQ01269623 GDIQ01267299 GDIQ01199098 GDIQ01184353 GDIQ01178604 GDIQ01165219 GDIQ01140626 GDIQ01129878 GDIQ01036152 JAL11100.1 GDIP01185927 JAJ37475.1 GDIQ01196295 JAK55430.1 GDIP01086691 JAM17024.1 GDIP01107393 JAL96321.1 GDIQ01123629 JAL28097.1 GDIP01157679 JAJ65723.1 GDIQ01009066 JAN85671.1 GDIQ01092283 JAL59443.1 GL732551 EFX79631.1 GDIP01018111 JAM85604.1 GDIQ01029761 JAN64976.1 GDIP01107394 JAL96320.1 NCKU01006687 RWS03249.1 GDIQ01169856 JAK81869.1 NEDP02005302 OWF42253.1 FN598566 CBI63263.1 EU287986 ABZ79726.1 MRZV01000410 PIK50612.1 BEZZ01000418 GCC32179.1 GL193258 EFB19059.1 BFAA01003941 GCB62814.1 AAQR03070941 AAQR03070942 AAQR03070943 GABZ01000800 JAA52725.1 ACTA01083799 AHAT01005284 AHAT01005285 AHAT01005286 AHAT01005287
ODYU01002714 SOQ40567.1 NWSH01000207 PCG78420.1 PCG78419.1 PCG78418.1 JTDY01000502 KOB76873.1 KQ459582 KPI99014.1 NEVH01021221 PNF19756.1 KK853093 KDR11510.1 PYGN01000219 PSN50906.1 PNF19757.1 PNF19755.1 GEDC01008120 JAS29178.1 GBYB01000919 JAG70686.1 GEBQ01005597 JAT34380.1 KQ971347 EFA05489.1 GEDC01013338 JAS23960.1 NNAY01001310 OXU24398.1 KZ288254 PBC30888.1 GEZM01007055 JAV95444.1 GEZM01007054 JAV95445.1 KQ767320 OAD53411.1 DS235171 EEB12945.1 KQ434972 KZC12738.1 GL762647 EFZ20995.1 KK107078 QOIP01000011 EZA60358.1 RLU16920.1 GL441701 EFN64407.1 ADTU01013171 KQ976401 KYM92587.1 GL449036 EFN83423.1 KQ981335 KYN42644.1 LBMM01000361 KMQ99399.1 KB632330 ERL92570.1 KQ982769 KYQ50870.1 GL888182 EGI65726.1 APGK01055716 APGK01055717 KB741269 ENN71457.1 GGMS01010776 MBY79979.1 ABLF02033059 ABLF02033063 LJIG01000129 KRT86811.1 GECU01018154 JAS89552.1 GDIQ01166711 JAK85014.1 GDIP01092541 JAM11174.1 GDIQ01081934 JAN12803.1 GDIQ01191592 JAK60133.1 GDIP01203162 JAJ20240.1 GDIP01207313 JAJ16089.1 GDIP01122333 JAL81381.1 GDIP01235565 JAI87836.1 GDIP01181671 JAJ41731.1 GDIQ01253826 GDIQ01252532 GDIQ01251187 GDIQ01208203 GDIQ01206598 GDIQ01205148 GDIP01091575 LRGB01000944 JAK43522.1 JAM12140.1 KZS14480.1 GDIP01216023 JAJ07379.1 GDIP01218171 JAJ05231.1 GDIP01083883 JAM19832.1 GDIQ01253827 GDIQ01252533 GDIQ01251188 GDIQ01248042 GDIQ01208204 GDIQ01206599 GDIQ01205149 GDIQ01203451 JAJ97897.1 GDIQ01192649 GDIQ01094534 JAL57192.1 GDIQ01209199 GDIQ01201697 GDIQ01184354 GDIQ01084837 JAK67371.1 GDIQ01269623 GDIQ01267299 GDIQ01199098 GDIQ01184353 GDIQ01178604 GDIQ01165219 GDIQ01140626 GDIQ01129878 GDIQ01036152 JAL11100.1 GDIP01185927 JAJ37475.1 GDIQ01196295 JAK55430.1 GDIP01086691 JAM17024.1 GDIP01107393 JAL96321.1 GDIQ01123629 JAL28097.1 GDIP01157679 JAJ65723.1 GDIQ01009066 JAN85671.1 GDIQ01092283 JAL59443.1 GL732551 EFX79631.1 GDIP01018111 JAM85604.1 GDIQ01029761 JAN64976.1 GDIP01107394 JAL96320.1 NCKU01006687 RWS03249.1 GDIQ01169856 JAK81869.1 NEDP02005302 OWF42253.1 FN598566 CBI63263.1 EU287986 ABZ79726.1 MRZV01000410 PIK50612.1 BEZZ01000418 GCC32179.1 GL193258 EFB19059.1 BFAA01003941 GCB62814.1 AAQR03070941 AAQR03070942 AAQR03070943 GABZ01000800 JAA52725.1 ACTA01083799 AHAT01005284 AHAT01005285 AHAT01005286 AHAT01005287
Proteomes
UP000007151
UP000053240
UP000283053
UP000218220
UP000037510
UP000053268
+ More
UP000235965 UP000027135 UP000245037 UP000007266 UP000002358 UP000215335 UP000005203 UP000242457 UP000009046 UP000076502 UP000192223 UP000053097 UP000279307 UP000000311 UP000005205 UP000078540 UP000008237 UP000078541 UP000036403 UP000030742 UP000075809 UP000007755 UP000019118 UP000007819 UP000076858 UP000000305 UP000285301 UP000242188 UP000007646 UP000230750 UP000287033 UP000286641 UP000248481 UP000085678 UP000288216 UP000005225 UP000261520 UP000261640 UP000008912 UP000018468 UP000261681
UP000235965 UP000027135 UP000245037 UP000007266 UP000002358 UP000215335 UP000005203 UP000242457 UP000009046 UP000076502 UP000192223 UP000053097 UP000279307 UP000000311 UP000005205 UP000078540 UP000008237 UP000078541 UP000036403 UP000030742 UP000075809 UP000007755 UP000019118 UP000007819 UP000076858 UP000000305 UP000285301 UP000242188 UP000007646 UP000230750 UP000287033 UP000286641 UP000248481 UP000085678 UP000288216 UP000005225 UP000261520 UP000261640 UP000008912 UP000018468 UP000261681
Pfam
PF00520 Ion_trans
Interpro
SUPFAM
SSF47473
SSF47473
Gene 3D
ProteinModelPortal
A0A212FMX6
A0A194R2B4
A0A437BBZ4
A0A2H1VIA3
A0A2A4K419
A0A2A4K2Z5
+ More
A0A2A4K3C1 A0A0L7LMU2 A0A194Q1Z0 A0A2J7PTT9 A0A067QPX2 A0A2P8Z326 A0A2J7PTU8 A0A2J7PTU6 A0A1B6DU32 A0A0C9QCA4 A0A1B6MEL7 D2A6B3 A0A1B6DE94 K7IN16 A0A232F1S9 A0A088A8Q4 A0A2A3EGQ6 A0A1Y1NBW7 A0A1Y1NFP5 A0A310SA12 E0VHQ9 A0A154PN53 E9IEK8 A0A1W4WI41 A0A026WWZ4 E2AQ73 A0A1W4WSY0 A0A158NEU4 A0A195BVH7 E2BLT9 A0A195FRV6 A0A0J7L7X6 U4UR78 A0A151WSM0 F4WJE8 N6TYV1 A0A2S2QQC8 J9K983 A0A0T6BI11 A0A1B6IRK8 A0A0P5LQC5 A0A0P5W0M2 A0A0P6DAA0 A0A0P5K371 A0A0P5A761 A0A0N7ZVX8 A0A0P5TS73 A0A0P4XWY9 A0A0P5BRX1 A0A0N8B6F7 A0A0P5A5V7 A0A0P4ZEA0 A0A0P5WQ39 A0A0N8AQ54 A0A0P5S0P0 A0A0P5M5U4 A0A0P5NLE8 A0A0P5BDU6 A0A0P5LE06 A0A0P5WGR0 A0A0P5V3X6 A0A0P5QI87 A0A0P5DE72 A0A0P6I5P3 A0A0N8CCU7 E9GLT0 A0A0N8DMW7 A0A0P6GTU1 A0A0P5UXL4 A0A3S3Q4H4 A0A0P5LGN1 A0A210Q0H2 D1J6X7 G3T3S0 G3TRD5 B8PXJ8 A0A2G8KRI8 A0A401SP53 A0A3Q7QIU4 A0A2Y9HY96 D2HS01 A0A1S3H6H9 A0A401NPM2 H0XD96 A0A3B4B558 K9IU16 A0A3Q7RXX2 A0A3Q3M710 G1LQ73 W5MPH5 A0A383Z701 A0A383Z7D9 A0A2Y9EMS6
A0A2A4K3C1 A0A0L7LMU2 A0A194Q1Z0 A0A2J7PTT9 A0A067QPX2 A0A2P8Z326 A0A2J7PTU8 A0A2J7PTU6 A0A1B6DU32 A0A0C9QCA4 A0A1B6MEL7 D2A6B3 A0A1B6DE94 K7IN16 A0A232F1S9 A0A088A8Q4 A0A2A3EGQ6 A0A1Y1NBW7 A0A1Y1NFP5 A0A310SA12 E0VHQ9 A0A154PN53 E9IEK8 A0A1W4WI41 A0A026WWZ4 E2AQ73 A0A1W4WSY0 A0A158NEU4 A0A195BVH7 E2BLT9 A0A195FRV6 A0A0J7L7X6 U4UR78 A0A151WSM0 F4WJE8 N6TYV1 A0A2S2QQC8 J9K983 A0A0T6BI11 A0A1B6IRK8 A0A0P5LQC5 A0A0P5W0M2 A0A0P6DAA0 A0A0P5K371 A0A0P5A761 A0A0N7ZVX8 A0A0P5TS73 A0A0P4XWY9 A0A0P5BRX1 A0A0N8B6F7 A0A0P5A5V7 A0A0P4ZEA0 A0A0P5WQ39 A0A0N8AQ54 A0A0P5S0P0 A0A0P5M5U4 A0A0P5NLE8 A0A0P5BDU6 A0A0P5LE06 A0A0P5WGR0 A0A0P5V3X6 A0A0P5QI87 A0A0P5DE72 A0A0P6I5P3 A0A0N8CCU7 E9GLT0 A0A0N8DMW7 A0A0P6GTU1 A0A0P5UXL4 A0A3S3Q4H4 A0A0P5LGN1 A0A210Q0H2 D1J6X7 G3T3S0 G3TRD5 B8PXJ8 A0A2G8KRI8 A0A401SP53 A0A3Q7QIU4 A0A2Y9HY96 D2HS01 A0A1S3H6H9 A0A401NPM2 H0XD96 A0A3B4B558 K9IU16 A0A3Q7RXX2 A0A3Q3M710 G1LQ73 W5MPH5 A0A383Z701 A0A383Z7D9 A0A2Y9EMS6
PDB
6C9A
E-value=1.30419e-99,
Score=927
Ontologies
GO
PANTHER
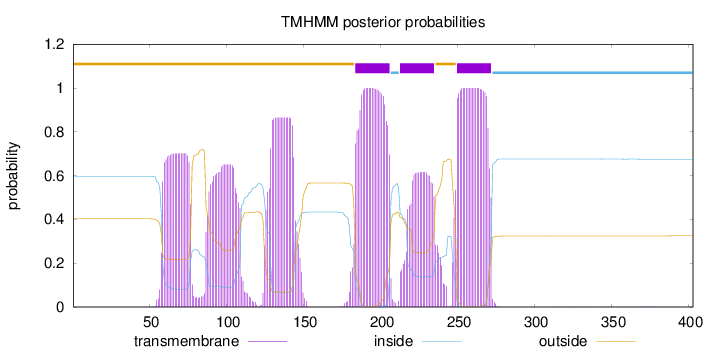
Topology
Length:
403
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
102.81057
Exp number, first 60 AAs:
1.63716
Total prob of N-in:
0.59710
outside
1 - 183
TMhelix
184 - 206
inside
207 - 212
TMhelix
213 - 235
outside
236 - 249
TMhelix
250 - 272
inside
273 - 403
Population Genetic Test Statistics
Pi
256.668416
Theta
166.057567
Tajima's D
1.497146
CLR
0.163929
CSRT
0.786210689465527
Interpretation
Uncertain
