Pre Gene Modal
BGIBMGA012783
Annotation
PREDICTED:_uncharacterized_protein_LOC105842042_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.038
Sequence
CDS
ATGAAAACTGCTTATCAAGTTTACGAGTATTTAGAAGGGGATGAAGATGACGGCTCAAAAATAGATTGGGAGGAATTAACTAAATTAACTGTATTAAATCATGTAATAGAACAACAAGAAAGAGATCTATTGCTCCAGACAGCTTTTTCGGAACAAAAACCAATTACAAGTAAAAGTGAAACGGCTCTTGAGAGAAAAAATAGCGAATTTGAACGGTTAGAGAAGAGATTCAGCTGTAGTGAAATTGAAGGCAAATCTAGACGACGATCAATACCTCAGGAAGCAAGAGTAGAGATGCTTGGTGTCTGGACTGAAGCAAACTTGAATTGTAAATTAAATGACGTAATAAACGAAGGAATTTTGGATTCTATACTTCCATATTTAGTTGGTTATAAAAAGCCTTCAAAGACGACCAGTGTTTATGTGCCGGTTATTAAAAAAACACCGTCTAATCCTACTACGGATGTGAAAAAACCAGCCAGTTTTGGAGGGTTTATAAATGAAAAAGACTCAAGAGATCGTCTCGGAAGGCGTAAATCATCTGTAATTGCAGCTCAAGACCGCGCGAATCATAAACAAGATGGCGACGTTGAAATCCACGTCTGCGATGAAGTCAAAGGCCTCAAGAAAGATTTTAGATGCCCACAAAAATTATTGGTTTCCAAAATGGGTTACTTTGCTGACGTGACCGCGGGTCAGAGACTTGAAGATATGGATATATCTGTTCACTGCGATATACAAATCTTCGATTGGTTGATGCGTTGGATGAAGAAGGATTCTATTTTAGTTGCGGATTGGCCCTTGTTGGATCCCCACAATGTGGTCCCTATACTGGTCTCGGCGTCTTTCTTACAGATGGAGCCATTGTTGCACGATTGTTTGATCTACTGTCATGCCCACATGAATGACATTGTCAAGACCACCACGAATCTGGCGTGTTTGAGCGACGCTTTACTGACAAGGCAAGTATTTGTTTCTATCTTATTACATAAAAAGTAG
Protein
MKTAYQVYEYLEGDEDDGSKIDWEELTKLTVLNHVIEQQERDLLLQTAFSEQKPITSKSETALERKNSEFERLEKRFSCSEIEGKSRRRSIPQEARVEMLGVWTEANLNCKLNDVINEGILDSILPYLVGYKKPSKTTSVYVPVIKKTPSNPTTDVKKPASFGGFINEKDSRDRLGRRKSSVIAAQDRANHKQDGDVEIHVCDEVKGLKKDFRCPQKLLVSKMGYFADVTAGQRLEDMDISVHCDIQIFDWLMRWMKKDSILVADWPLLDPHNVVPILVSASFLQMEPLLHDCLIYCHAHMNDIVKTTTNLACLSDALLTRQVFVSILLHKK
Summary
Uniprot
H9JTC1
A0A2A4K323
A0A212FMX9
A0A194R1T6
A0A194Q0A5
A0A2H1W732
+ More
A0A0L7KNR0 A0A1Q3FT16 B0WC61 A0A1B0GJ25 A0A1S4F6V2 A0A182JVK8 A0A182UF76 A0A182JCH6 Q0IFU1 A0A182T3V5 A0A1B0D410 A0A182LI14 Q5TSK3 A0A182MLW1 A0A182XNM2 A0A336LZB2 A0A182HXP8 A0A182PSZ8 A0A182NNI9 A0A151XGK5 A0A182RKJ9 A0A084VXI1 A0A182WCT7 A0A182Q0T7 A0A336MZE8 A0A182YPI4 A0A0C9R8S9 A0A232EZP0 A0A151JV19 A0A026W4C0 A0A1J1I9G3 A0A0L7RES3 A0A1Y1JXF8 A0A0J7L5F9 A0A0L0CDA3 E9IW38 A0A1I8PIX5 B4KVV1 A0A1I8N6Z8 A0A1B0B5I6 A0A1A9Y0R7 A0A0M4EGT0 A0A1A9V3N4 A0A1B0G8F5 A0A3L8D4G0 B4H7Z6 Q29D48 A0A0R3P7L7 Q9VT34 Q95U50 M9PEY8 A0A1B0AI37 A0A0A1X3C9 A0A0A1XNV6 A0A0K8V3N0 A0A034UYV7 A0A1A9WTM3
A0A0L7KNR0 A0A1Q3FT16 B0WC61 A0A1B0GJ25 A0A1S4F6V2 A0A182JVK8 A0A182UF76 A0A182JCH6 Q0IFU1 A0A182T3V5 A0A1B0D410 A0A182LI14 Q5TSK3 A0A182MLW1 A0A182XNM2 A0A336LZB2 A0A182HXP8 A0A182PSZ8 A0A182NNI9 A0A151XGK5 A0A182RKJ9 A0A084VXI1 A0A182WCT7 A0A182Q0T7 A0A336MZE8 A0A182YPI4 A0A0C9R8S9 A0A232EZP0 A0A151JV19 A0A026W4C0 A0A1J1I9G3 A0A0L7RES3 A0A1Y1JXF8 A0A0J7L5F9 A0A0L0CDA3 E9IW38 A0A1I8PIX5 B4KVV1 A0A1I8N6Z8 A0A1B0B5I6 A0A1A9Y0R7 A0A0M4EGT0 A0A1A9V3N4 A0A1B0G8F5 A0A3L8D4G0 B4H7Z6 Q29D48 A0A0R3P7L7 Q9VT34 Q95U50 M9PEY8 A0A1B0AI37 A0A0A1X3C9 A0A0A1XNV6 A0A0K8V3N0 A0A034UYV7 A0A1A9WTM3
Pubmed
EMBL
BABH01004154
NWSH01000207
PCG78416.1
AGBW02007651
OWR55104.1
KQ460878
+ More
KPJ11657.1 KQ459582 KPI99012.1 ODYU01006755 SOQ48891.1 JTDY01008108 KOB64735.1 GFDL01004328 JAV30717.1 DS231885 EDS43213.1 AJWK01019227 CH477294 EAT44441.1 AJVK01023857 AJVK01023858 AJVK01023859 AJVK01023860 AAAB01008905 EAL40403.3 AXCM01001770 UFQT01000344 SSX23416.1 APCN01005335 KQ982169 KYQ59441.1 ATLV01018074 KE525212 KFB42675.1 AXCN02001913 UFQS01001638 UFQT01001638 SSX11713.1 SSX31278.1 GBYB01004560 JAG74327.1 NNAY01001458 OXU23904.1 KQ981713 KYN37367.1 KK107429 EZA50930.1 CVRI01000043 CRK96212.1 KQ414609 KOC69330.1 GEZM01098108 JAV53989.1 LBMM01000694 KMQ97724.1 JRES01000534 KNC30373.1 GL766449 EFZ15274.1 CH933809 EDW18475.2 JXJN01008733 CP012525 ALC43677.1 CCAG010000249 QOIP01000014 RLU14718.1 CH479219 EDW34786.1 CH379070 EAL30566.3 KRT09054.1 AE014296 AAF50221.1 AY058308 AAL13537.1 AGB94340.1 GBXI01008493 JAD05799.1 GBXI01001722 JAD12570.1 GDHF01018858 JAI33456.1 GAKP01022786 JAC36166.1
KPJ11657.1 KQ459582 KPI99012.1 ODYU01006755 SOQ48891.1 JTDY01008108 KOB64735.1 GFDL01004328 JAV30717.1 DS231885 EDS43213.1 AJWK01019227 CH477294 EAT44441.1 AJVK01023857 AJVK01023858 AJVK01023859 AJVK01023860 AAAB01008905 EAL40403.3 AXCM01001770 UFQT01000344 SSX23416.1 APCN01005335 KQ982169 KYQ59441.1 ATLV01018074 KE525212 KFB42675.1 AXCN02001913 UFQS01001638 UFQT01001638 SSX11713.1 SSX31278.1 GBYB01004560 JAG74327.1 NNAY01001458 OXU23904.1 KQ981713 KYN37367.1 KK107429 EZA50930.1 CVRI01000043 CRK96212.1 KQ414609 KOC69330.1 GEZM01098108 JAV53989.1 LBMM01000694 KMQ97724.1 JRES01000534 KNC30373.1 GL766449 EFZ15274.1 CH933809 EDW18475.2 JXJN01008733 CP012525 ALC43677.1 CCAG010000249 QOIP01000014 RLU14718.1 CH479219 EDW34786.1 CH379070 EAL30566.3 KRT09054.1 AE014296 AAF50221.1 AY058308 AAL13537.1 AGB94340.1 GBXI01008493 JAD05799.1 GBXI01001722 JAD12570.1 GDHF01018858 JAI33456.1 GAKP01022786 JAC36166.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000037510
+ More
UP000002320 UP000092461 UP000075881 UP000075902 UP000075880 UP000008820 UP000075901 UP000092462 UP000075882 UP000007062 UP000075883 UP000076407 UP000075840 UP000075885 UP000075884 UP000075809 UP000075900 UP000030765 UP000075920 UP000075886 UP000076408 UP000215335 UP000078541 UP000053097 UP000183832 UP000053825 UP000036403 UP000037069 UP000095300 UP000009192 UP000095301 UP000092460 UP000092443 UP000092553 UP000078200 UP000092444 UP000279307 UP000008744 UP000001819 UP000000803 UP000092445 UP000091820
UP000002320 UP000092461 UP000075881 UP000075902 UP000075880 UP000008820 UP000075901 UP000092462 UP000075882 UP000007062 UP000075883 UP000076407 UP000075840 UP000075885 UP000075884 UP000075809 UP000075900 UP000030765 UP000075920 UP000075886 UP000076408 UP000215335 UP000078541 UP000053097 UP000183832 UP000053825 UP000036403 UP000037069 UP000095300 UP000009192 UP000095301 UP000092460 UP000092443 UP000092553 UP000078200 UP000092444 UP000279307 UP000008744 UP000001819 UP000000803 UP000092445 UP000091820
Pfam
PF11822 DUF3342
SUPFAM
SSF54695
SSF54695
ProteinModelPortal
H9JTC1
A0A2A4K323
A0A212FMX9
A0A194R1T6
A0A194Q0A5
A0A2H1W732
+ More
A0A0L7KNR0 A0A1Q3FT16 B0WC61 A0A1B0GJ25 A0A1S4F6V2 A0A182JVK8 A0A182UF76 A0A182JCH6 Q0IFU1 A0A182T3V5 A0A1B0D410 A0A182LI14 Q5TSK3 A0A182MLW1 A0A182XNM2 A0A336LZB2 A0A182HXP8 A0A182PSZ8 A0A182NNI9 A0A151XGK5 A0A182RKJ9 A0A084VXI1 A0A182WCT7 A0A182Q0T7 A0A336MZE8 A0A182YPI4 A0A0C9R8S9 A0A232EZP0 A0A151JV19 A0A026W4C0 A0A1J1I9G3 A0A0L7RES3 A0A1Y1JXF8 A0A0J7L5F9 A0A0L0CDA3 E9IW38 A0A1I8PIX5 B4KVV1 A0A1I8N6Z8 A0A1B0B5I6 A0A1A9Y0R7 A0A0M4EGT0 A0A1A9V3N4 A0A1B0G8F5 A0A3L8D4G0 B4H7Z6 Q29D48 A0A0R3P7L7 Q9VT34 Q95U50 M9PEY8 A0A1B0AI37 A0A0A1X3C9 A0A0A1XNV6 A0A0K8V3N0 A0A034UYV7 A0A1A9WTM3
A0A0L7KNR0 A0A1Q3FT16 B0WC61 A0A1B0GJ25 A0A1S4F6V2 A0A182JVK8 A0A182UF76 A0A182JCH6 Q0IFU1 A0A182T3V5 A0A1B0D410 A0A182LI14 Q5TSK3 A0A182MLW1 A0A182XNM2 A0A336LZB2 A0A182HXP8 A0A182PSZ8 A0A182NNI9 A0A151XGK5 A0A182RKJ9 A0A084VXI1 A0A182WCT7 A0A182Q0T7 A0A336MZE8 A0A182YPI4 A0A0C9R8S9 A0A232EZP0 A0A151JV19 A0A026W4C0 A0A1J1I9G3 A0A0L7RES3 A0A1Y1JXF8 A0A0J7L5F9 A0A0L0CDA3 E9IW38 A0A1I8PIX5 B4KVV1 A0A1I8N6Z8 A0A1B0B5I6 A0A1A9Y0R7 A0A0M4EGT0 A0A1A9V3N4 A0A1B0G8F5 A0A3L8D4G0 B4H7Z6 Q29D48 A0A0R3P7L7 Q9VT34 Q95U50 M9PEY8 A0A1B0AI37 A0A0A1X3C9 A0A0A1XNV6 A0A0K8V3N0 A0A034UYV7 A0A1A9WTM3
PDB
4YDH
E-value=0.0251613,
Score=87
Ontologies
GO
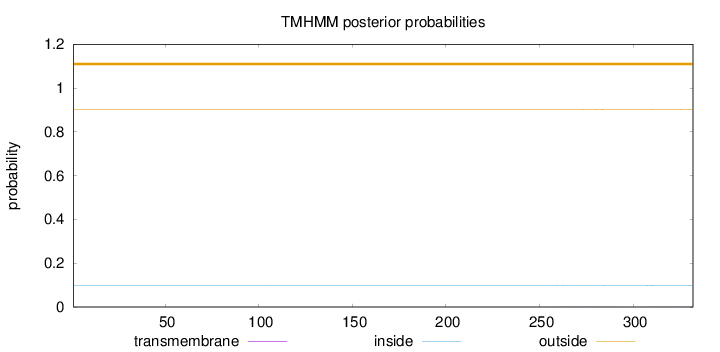
Topology
Length:
332
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05106
Exp number, first 60 AAs:
0
Total prob of N-in:
0.09866
outside
1 - 332
Population Genetic Test Statistics
Pi
275.79758
Theta
154.094822
Tajima's D
0.487529
CLR
0.339035
CSRT
0.505474726263687
Interpretation
Uncertain
