Pre Gene Modal
BGIBMGA013077
Annotation
PREDICTED:_TBC1_domain_family_member_22B_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.646 Nuclear Reliability : 1.285
Sequence
CDS
ATGGAAAATCACGAACCCGAACATGACAAGGGTTTGTTGTCTGACACGTTCTGGAGGACACCTCAGCGCGCTGTACCGGGGAGGCCTTACCAAAGGGTCATCCCAAACGAGGTTATGTCCGGGTCGTCGTATCAGCAATTCCAGGCGTCAGTCAACGACGCCTGGGATCTCGGCGACGACGAGATCATCTCGGGGATAGCGGACACTAAAATATCGAAAAGTATATCGGAAACAGCAGCCTTGAACGTCATAAATTCCCACAGAAACAGCACTAAAAACGTAAACAGAAGTGACGTGAAGGTTGAGAGTGAAGTTAAGAGTAAAGAGGTGGTGGAAGTTAAGAAGAATGAGGGTCGGAAGGTCGGTGGTGGGTCGGGTATAAGGCAGTATCCGGGTCGGCCTCGCCCAGTGCGGCTATCAGCGCTCGCTTCACGCGAGGAGAGCAGTGAGTCTAAATTGGAGCGCTTCCAGCAGATACACGATAGCACAGTCGTCAACCTCGACGAACTCAAACAGATCAGCTGGTCTGGGATACCAGTTAAAGCTAGAGCTGTAACATGGAGGCTTCTGGCCGGCTACTTACCAGCTAATTCTGAACGTCGCGCAGACACACTTCAACGAAAACGTGCCGATTACAAACATCTAGTAAGACAATATTATGATGCAGAAAGAGACGATGATACTTACCGACAGATACATATTGATATACCAAGAATGAGCCCACTCGTAGCTCTGTTCCAACAGAACACTGTGCAGGTTATGTTCGAGAGGATCTTGTATATTTGGGCAATAAGACACCCAGCTTCCGGCTACGTCCAAGGCATTAATGACCTCGTGACGCCGTTCTTCATGGTGTTCCTCCAGGAGGCTGTTCCTGGCGAGGAGCTGGATAATTATCCTCTGGACAAGTTGACTGAAGAGAAGAGGGACATCATCGAGGCAGACTCGTTTTGGTGTCTCTCGAAATTCTTAGACAGCGTTCAGGATAACTATATTTTTGCACAGCTGGGAATACAATACAAGGTAAATCAATTGAAGGAGCTGATAAGACGCATCGATCTACCACTCCACGAGCATTTGCAGCAGCACGGTGTGGACTACCTCCAGTTCTCATTCCGCTGGATGAACAATCTCCTCACGAGAGAAATACCGTTAGCCTGTACCATTCGATTATGGGACACTTATCTTGCAGAATCCGACGAGTTCGCTACATTTCAACTCTACGTCTGTGCCGCTTTCCTGCTTCACTGGAGAGAGAGATTGATGCTGGAAAGAGATTTCCAAGGTCTAATGATATTGTTACAGAACGTACCAACACAAAATTGGACTGATTCAAATATAAGTGTACTAGTTGCAGAGGCGTATAGGCTTAAGTTTGCTTTTGCCGATGCCCCCAAACATCTGCAGAGCGGTAAAGCAGACAGGTGA
Protein
MENHEPEHDKGLLSDTFWRTPQRAVPGRPYQRVIPNEVMSGSSYQQFQASVNDAWDLGDDEIISGIADTKISKSISETAALNVINSHRNSTKNVNRSDVKVESEVKSKEVVEVKKNEGRKVGGGSGIRQYPGRPRPVRLSALASREESSESKLERFQQIHDSTVVNLDELKQISWSGIPVKARAVTWRLLAGYLPANSERRADTLQRKRADYKHLVRQYYDAERDDDTYRQIHIDIPRMSPLVALFQQNTVQVMFERILYIWAIRHPASGYVQGINDLVTPFFMVFLQEAVPGEELDNYPLDKLTEEKRDIIEADSFWCLSKFLDSVQDNYIFAQLGIQYKVNQLKELIRRIDLPLHEHLQQHGVDYLQFSFRWMNNLLTREIPLACTIRLWDTYLAESDEFATFQLYVCAAFLLHWRERLMLERDFQGLMILLQNVPTQNWTDSNISVLVAEAYRLKFAFADAPKHLQSGKADR
Summary
Uniprot
H9JU65
A0A2A4K3R3
A0A437BC27
A0A194R712
A0A194Q0C7
A0A212FMY6
+ More
A0A2J7RSU5 A0A026W1P9 A0A1B6D9K2 K7IXR5 A0A0C9RC24 A0A3L8D678 A0A182K0F4 Q7Q9Y8 A0A182SZ38 A0A232EIQ5 A0A182XIG3 A0A182RE18 A0A1W4WE70 A0A088AUX0 A0A1W4WP62 A0A2H1W740 A0A2J7RSV6 A0A182H9N3 A0A1Y1NF45 A0A2A3E645 A0A195D1Y1 A0A182N9X4 A0A336M303 A0A336K258 B4PQA6 A0A3B0JME3 A0A182GG72 A0A0K8TMD8 A0A1W4VY72 B4R148 B4IKL1 Q95RC5 A0A1Q3FAN7 A0A1Q3FCU9 B3P2Y4 B3M2W3 A0A1Q3FD89 Q296V3 B4KDM0 A0A1L8DVM4 A0A1L8DW72 A0A0M4EDB5 J9JT79 A0A195F3Q6 E9IKP8 A0A151WXX4 B4JFJ1 A0A158NZQ4 A0A151I3R1 A0A0M9A0Q8 A0A151IXW8 D6W7S7 E2AAW3 A0A154PMU2 A0A2M4BNZ7 A0A2M4AHS6 U4TX30 A0A2M3Z7H4 W8BWT6 A0A310SVT0 B4LWV2 A0A2M4BJ23 W5JIL0 N6U6L5 A0A1J1I1V2 A0A182PHW3 A0A1Y9GL57 A0A1S4GL92 A0A0P8YND2 A0A182Y8Q4 E0VAT9 Q16QC4 A0A034W8K0 B4NH53 A0A0K8V520 A0A084WTY0 A0A182VEG5 A0A1S4FSU4 A0A1A9W9E3 A0A182KZH5 A0A0L7QLA4 E2C0B1 F4WRJ9 A0A182QNE4 T1PHV4 A0A165A4Z5 A0A0P5Z6F9 A0A2R5LCT5 A0A182W3I4
A0A2J7RSU5 A0A026W1P9 A0A1B6D9K2 K7IXR5 A0A0C9RC24 A0A3L8D678 A0A182K0F4 Q7Q9Y8 A0A182SZ38 A0A232EIQ5 A0A182XIG3 A0A182RE18 A0A1W4WE70 A0A088AUX0 A0A1W4WP62 A0A2H1W740 A0A2J7RSV6 A0A182H9N3 A0A1Y1NF45 A0A2A3E645 A0A195D1Y1 A0A182N9X4 A0A336M303 A0A336K258 B4PQA6 A0A3B0JME3 A0A182GG72 A0A0K8TMD8 A0A1W4VY72 B4R148 B4IKL1 Q95RC5 A0A1Q3FAN7 A0A1Q3FCU9 B3P2Y4 B3M2W3 A0A1Q3FD89 Q296V3 B4KDM0 A0A1L8DVM4 A0A1L8DW72 A0A0M4EDB5 J9JT79 A0A195F3Q6 E9IKP8 A0A151WXX4 B4JFJ1 A0A158NZQ4 A0A151I3R1 A0A0M9A0Q8 A0A151IXW8 D6W7S7 E2AAW3 A0A154PMU2 A0A2M4BNZ7 A0A2M4AHS6 U4TX30 A0A2M3Z7H4 W8BWT6 A0A310SVT0 B4LWV2 A0A2M4BJ23 W5JIL0 N6U6L5 A0A1J1I1V2 A0A182PHW3 A0A1Y9GL57 A0A1S4GL92 A0A0P8YND2 A0A182Y8Q4 E0VAT9 Q16QC4 A0A034W8K0 B4NH53 A0A0K8V520 A0A084WTY0 A0A182VEG5 A0A1S4FSU4 A0A1A9W9E3 A0A182KZH5 A0A0L7QLA4 E2C0B1 F4WRJ9 A0A182QNE4 T1PHV4 A0A165A4Z5 A0A0P5Z6F9 A0A2R5LCT5 A0A182W3I4
Pubmed
19121390
26354079
22118469
24508170
20075255
30249741
+ More
12364791 28648823 26483478 28004739 17994087 17550304 26369729 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 21282665 21347285 18362917 19820115 20798317 23537049 24495485 20920257 23761445 25244985 20566863 17510324 25348373 24438588 20966253 21719571
12364791 28648823 26483478 28004739 17994087 17550304 26369729 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 21282665 21347285 18362917 19820115 20798317 23537049 24495485 20920257 23761445 25244985 20566863 17510324 25348373 24438588 20966253 21719571
EMBL
BABH01004154
BABH01004155
NWSH01000207
PCG78403.1
RSAL01000093
RVE47918.1
+ More
KQ460878 KPJ11656.1 KQ459582 KPI99011.1 AGBW02007651 OWR55105.1 NEVH01000250 PNF43906.1 KK107488 EZA49995.1 GEDC01014956 JAS22342.1 GBYB01005805 JAG75572.1 QOIP01000012 RLU15965.1 AAAB01008898 EAA09194.4 NNAY01004198 OXU18212.1 ODYU01006755 SOQ48890.1 PNF43911.1 JXUM01121255 JXUM01121256 KQ566479 KXJ70100.1 GEZM01003859 JAV96522.1 KZ288356 PBC27178.1 KQ976948 KYN06913.1 UFQT01000478 SSX24646.1 UFQS01000059 UFQT01000059 SSW98769.1 SSX19155.1 CM000160 EDW96215.1 OUUW01000007 SPP83405.1 JXUM01011362 JXUM01011363 KQ560329 KXJ83008.1 GDAI01002285 JAI15318.1 CM000364 EDX12135.1 CH480853 EDW51615.1 AE014297 AY061479 AAF55847.2 AAL29027.1 GFDL01010438 JAV24607.1 GFDL01009640 JAV25405.1 CH954181 EDV48298.1 CH902617 EDV43493.1 GFDL01009491 JAV25554.1 CM000070 EAL28354.2 CH933806 EDW13854.1 GFDF01003615 JAV10469.1 GFDF01003589 JAV10495.1 CP012526 ALC45766.1 ABLF02027997 KQ981856 KYN34714.1 GL764026 EFZ18742.1 KQ982656 KYQ52728.1 CH916369 EDV93472.1 ADTU01004890 KQ976474 KYM83952.1 KQ435794 KOX73996.1 KQ980791 KYN13146.1 KQ971307 EFA11341.1 GL438191 EFN69426.1 KQ434954 KZC12618.1 GGFJ01005658 MBW54799.1 GGFK01007009 MBW40330.1 KB631792 ERL86169.1 GGFM01003704 MBW24455.1 GAMC01012883 GAMC01012881 JAB93674.1 KQ759866 OAD62483.1 CH940650 EDW66673.2 GGFJ01003863 MBW53004.1 ADMH02001067 ETN64222.1 APGK01047183 KB741077 ENN74202.1 CVRI01000026 CRK92353.1 APCN01005402 KPU80289.1 DS235015 EEB10494.1 CH477752 EAT36604.1 GAKP01007918 GAKP01007917 JAC51034.1 CH964272 EDW84550.2 GDHF01018338 JAI33976.1 ATLV01026943 KE525420 KFB53674.1 KQ414934 KOC59339.1 GL451753 EFN78663.1 GL888291 EGI63144.1 AXCN02001202 KA648269 AFP62898.1 LRGB01000642 KZS17184.1 GDIP01060955 JAM42760.1 GGLE01003153 MBY07279.1
KQ460878 KPJ11656.1 KQ459582 KPI99011.1 AGBW02007651 OWR55105.1 NEVH01000250 PNF43906.1 KK107488 EZA49995.1 GEDC01014956 JAS22342.1 GBYB01005805 JAG75572.1 QOIP01000012 RLU15965.1 AAAB01008898 EAA09194.4 NNAY01004198 OXU18212.1 ODYU01006755 SOQ48890.1 PNF43911.1 JXUM01121255 JXUM01121256 KQ566479 KXJ70100.1 GEZM01003859 JAV96522.1 KZ288356 PBC27178.1 KQ976948 KYN06913.1 UFQT01000478 SSX24646.1 UFQS01000059 UFQT01000059 SSW98769.1 SSX19155.1 CM000160 EDW96215.1 OUUW01000007 SPP83405.1 JXUM01011362 JXUM01011363 KQ560329 KXJ83008.1 GDAI01002285 JAI15318.1 CM000364 EDX12135.1 CH480853 EDW51615.1 AE014297 AY061479 AAF55847.2 AAL29027.1 GFDL01010438 JAV24607.1 GFDL01009640 JAV25405.1 CH954181 EDV48298.1 CH902617 EDV43493.1 GFDL01009491 JAV25554.1 CM000070 EAL28354.2 CH933806 EDW13854.1 GFDF01003615 JAV10469.1 GFDF01003589 JAV10495.1 CP012526 ALC45766.1 ABLF02027997 KQ981856 KYN34714.1 GL764026 EFZ18742.1 KQ982656 KYQ52728.1 CH916369 EDV93472.1 ADTU01004890 KQ976474 KYM83952.1 KQ435794 KOX73996.1 KQ980791 KYN13146.1 KQ971307 EFA11341.1 GL438191 EFN69426.1 KQ434954 KZC12618.1 GGFJ01005658 MBW54799.1 GGFK01007009 MBW40330.1 KB631792 ERL86169.1 GGFM01003704 MBW24455.1 GAMC01012883 GAMC01012881 JAB93674.1 KQ759866 OAD62483.1 CH940650 EDW66673.2 GGFJ01003863 MBW53004.1 ADMH02001067 ETN64222.1 APGK01047183 KB741077 ENN74202.1 CVRI01000026 CRK92353.1 APCN01005402 KPU80289.1 DS235015 EEB10494.1 CH477752 EAT36604.1 GAKP01007918 GAKP01007917 JAC51034.1 CH964272 EDW84550.2 GDHF01018338 JAI33976.1 ATLV01026943 KE525420 KFB53674.1 KQ414934 KOC59339.1 GL451753 EFN78663.1 GL888291 EGI63144.1 AXCN02001202 KA648269 AFP62898.1 LRGB01000642 KZS17184.1 GDIP01060955 JAM42760.1 GGLE01003153 MBY07279.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000053268
UP000007151
+ More
UP000235965 UP000053097 UP000002358 UP000279307 UP000075881 UP000007062 UP000075901 UP000215335 UP000076407 UP000075900 UP000192223 UP000005203 UP000069940 UP000249989 UP000242457 UP000078542 UP000075884 UP000002282 UP000268350 UP000192221 UP000000304 UP000001292 UP000000803 UP000008711 UP000007801 UP000001819 UP000009192 UP000092553 UP000007819 UP000078541 UP000075809 UP000001070 UP000005205 UP000078540 UP000053105 UP000078492 UP000007266 UP000000311 UP000076502 UP000030742 UP000008792 UP000000673 UP000019118 UP000183832 UP000075885 UP000075840 UP000076408 UP000009046 UP000008820 UP000007798 UP000030765 UP000075903 UP000091820 UP000075882 UP000053825 UP000008237 UP000007755 UP000075886 UP000076858 UP000075920
UP000235965 UP000053097 UP000002358 UP000279307 UP000075881 UP000007062 UP000075901 UP000215335 UP000076407 UP000075900 UP000192223 UP000005203 UP000069940 UP000249989 UP000242457 UP000078542 UP000075884 UP000002282 UP000268350 UP000192221 UP000000304 UP000001292 UP000000803 UP000008711 UP000007801 UP000001819 UP000009192 UP000092553 UP000007819 UP000078541 UP000075809 UP000001070 UP000005205 UP000078540 UP000053105 UP000078492 UP000007266 UP000000311 UP000076502 UP000030742 UP000008792 UP000000673 UP000019118 UP000183832 UP000075885 UP000075840 UP000076408 UP000009046 UP000008820 UP000007798 UP000030765 UP000075903 UP000091820 UP000075882 UP000053825 UP000008237 UP000007755 UP000075886 UP000076858 UP000075920
Interpro
SUPFAM
SSF47923
SSF47923
ProteinModelPortal
H9JU65
A0A2A4K3R3
A0A437BC27
A0A194R712
A0A194Q0C7
A0A212FMY6
+ More
A0A2J7RSU5 A0A026W1P9 A0A1B6D9K2 K7IXR5 A0A0C9RC24 A0A3L8D678 A0A182K0F4 Q7Q9Y8 A0A182SZ38 A0A232EIQ5 A0A182XIG3 A0A182RE18 A0A1W4WE70 A0A088AUX0 A0A1W4WP62 A0A2H1W740 A0A2J7RSV6 A0A182H9N3 A0A1Y1NF45 A0A2A3E645 A0A195D1Y1 A0A182N9X4 A0A336M303 A0A336K258 B4PQA6 A0A3B0JME3 A0A182GG72 A0A0K8TMD8 A0A1W4VY72 B4R148 B4IKL1 Q95RC5 A0A1Q3FAN7 A0A1Q3FCU9 B3P2Y4 B3M2W3 A0A1Q3FD89 Q296V3 B4KDM0 A0A1L8DVM4 A0A1L8DW72 A0A0M4EDB5 J9JT79 A0A195F3Q6 E9IKP8 A0A151WXX4 B4JFJ1 A0A158NZQ4 A0A151I3R1 A0A0M9A0Q8 A0A151IXW8 D6W7S7 E2AAW3 A0A154PMU2 A0A2M4BNZ7 A0A2M4AHS6 U4TX30 A0A2M3Z7H4 W8BWT6 A0A310SVT0 B4LWV2 A0A2M4BJ23 W5JIL0 N6U6L5 A0A1J1I1V2 A0A182PHW3 A0A1Y9GL57 A0A1S4GL92 A0A0P8YND2 A0A182Y8Q4 E0VAT9 Q16QC4 A0A034W8K0 B4NH53 A0A0K8V520 A0A084WTY0 A0A182VEG5 A0A1S4FSU4 A0A1A9W9E3 A0A182KZH5 A0A0L7QLA4 E2C0B1 F4WRJ9 A0A182QNE4 T1PHV4 A0A165A4Z5 A0A0P5Z6F9 A0A2R5LCT5 A0A182W3I4
A0A2J7RSU5 A0A026W1P9 A0A1B6D9K2 K7IXR5 A0A0C9RC24 A0A3L8D678 A0A182K0F4 Q7Q9Y8 A0A182SZ38 A0A232EIQ5 A0A182XIG3 A0A182RE18 A0A1W4WE70 A0A088AUX0 A0A1W4WP62 A0A2H1W740 A0A2J7RSV6 A0A182H9N3 A0A1Y1NF45 A0A2A3E645 A0A195D1Y1 A0A182N9X4 A0A336M303 A0A336K258 B4PQA6 A0A3B0JME3 A0A182GG72 A0A0K8TMD8 A0A1W4VY72 B4R148 B4IKL1 Q95RC5 A0A1Q3FAN7 A0A1Q3FCU9 B3P2Y4 B3M2W3 A0A1Q3FD89 Q296V3 B4KDM0 A0A1L8DVM4 A0A1L8DW72 A0A0M4EDB5 J9JT79 A0A195F3Q6 E9IKP8 A0A151WXX4 B4JFJ1 A0A158NZQ4 A0A151I3R1 A0A0M9A0Q8 A0A151IXW8 D6W7S7 E2AAW3 A0A154PMU2 A0A2M4BNZ7 A0A2M4AHS6 U4TX30 A0A2M3Z7H4 W8BWT6 A0A310SVT0 B4LWV2 A0A2M4BJ23 W5JIL0 N6U6L5 A0A1J1I1V2 A0A182PHW3 A0A1Y9GL57 A0A1S4GL92 A0A0P8YND2 A0A182Y8Q4 E0VAT9 Q16QC4 A0A034W8K0 B4NH53 A0A0K8V520 A0A084WTY0 A0A182VEG5 A0A1S4FSU4 A0A1A9W9E3 A0A182KZH5 A0A0L7QLA4 E2C0B1 F4WRJ9 A0A182QNE4 T1PHV4 A0A165A4Z5 A0A0P5Z6F9 A0A2R5LCT5 A0A182W3I4
PDB
6D0S
E-value=7.08155e-112,
Score=1034
Ontologies
PANTHER
Topology
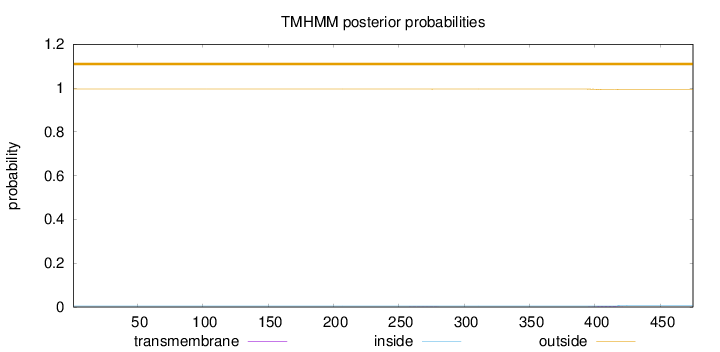
Length:
475
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0587800000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00478
outside
1 - 475
Population Genetic Test Statistics
Pi
243.088428
Theta
193.304242
Tajima's D
1.231419
CLR
0.653369
CSRT
0.720613969301535
Interpretation
Uncertain
