Gene
KWMTBOMO09433
Pre Gene Modal
BGIBMGA013079
Annotation
PREDICTED:_protein_strawberry_notch-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.607
Sequence
CDS
ATGTCAAGACGTGCCTACGCCGAGGATCCGGCGCCGTCAGATGATTCTGATTTCGATGATGAAGAGGATCCTGATAATTTGGAGGTGCCAGGTGGAGGTAAAACCCTGGCTGCTGCAGCCCGGATGGCCGACAAGAAGGTACCACAAGCAACTGCGCCACAAGTCACGGTGAGACCTACCACAAATCCACTTGCAGCACCGTCCGGCGTTGCGTTTGGACAAACGCACAACGGGAAGCCAATAAAGATACAGGCCAGCTCCTTGACGAAACCCTTCAATCTCCTCAACCTGCCAGGAGTCGGACGCGGAGGAATGGGCGCGGCGATGGCTCCGTTCGCGGCGGGTTCGTCGGCCGCATTCAACGCGGCCTCGTTACTCGCGCAGATGGAGATGGTCGGCATTACGGGCGTCAACTCGTATTTGCTGCAGAATCTCAATCAGATCATAGCGGCCGGCATGGGGGGCTCGATGCTCGGTCAGTTCTCGCAGCAGCTACCCCCCTGGGCTGGTCAGATCAAGCCCGGAATGTGGCACCAGGATAAGATGCCTGGAGAAGAAGAGGAAGTGGACGATGAAGAAATGGGAGTCGCAGAAACATATGCGGACTACATGCCGCCAAAGGTGAAACTAGGCAGGAAACATCCTGATCCTGTGGTGGAGACGGCGTCGTTGTCGTCCGTGGAGCCGGTGGACGTGACGTACAAGCTGAGTCTGCCGGACGAGACGATCCGGGGCGGGCTGCTGTCGGCCTTGCAGCTGGAGGCCGTGGTGTACGCGTCGCAGGCCCACGAGCACACGCTCCCGGACGGGACCAGGGCGGGCTTCCTCATAGGTGACGGTGCGGGTGTGGGCAAAGGTCGCACGATAGCCGGCATTATATTCGAGAACTACATCAAGGGTCGGAAGCGAGCCATATGGGTGTCCGTCTCCAACGACCTCAAGTACGACGCCGAGAGGGACCTCCGGGACATCGGGGCCGGGAAAATTGAAGTCTACCCTCTCAATAAGTTCAAGTACGCGAAGATCTCGTCGGCCGTGAACGGCAACGTGAAGAAGGGGGTCGTGTTCGCCACCTACTCCTCGCTCATCGGCGAGACCGCCAGCAACAACACCAAGTACAGGTCCAGGCTGAAGCAGCTGCTGCAGTGGTGCGGGGAGGACTTTGACGGAGTCATCGTATTTGACGAGTGTCACAAGGCAAAGAACCTCTGCCCCGTCGGCTCCGGGAAGGCGACGAAGACAGGTCTGACGGCCCTCGAGCTGCAGAACAAGCTGCCCAAGGCGAGGGTGGTGTACGCGTCCGCCACCGGGGCCTCGGAGCCCAGGAACATGGCGTACATGGTGCGCCTCGGGATATGGGGGGAGGGGACGCCCTTCCCGACCTTTATGGACTTCATTAACGCTGTGGAGAAGAGAGGCGTGGGCGCCATGGAGATAGTGGCGATGGACATGAAGCTGCGCGGGATGTACATAGCGCGCCAGCTGTCCTTCCACGGAGTGTCCTTCCGCATCGAGGAGGTGCCGCTCTCCGACGCCTTCAAGGAGACCTACGACAAGAGCGTCGCGCTGTGGGTGGAGGCCATGCAGCGGTTCACGGAGGCCGCCGAGCTGATAGACGCGGAGTCGAGGATGAGGAAGACCATGTGGGGACAGTTCTGGTCTGCTCATCAGAGGTTCTTCAAGTACCTGTGTATTGCCGCTAAGGTGAACCACGCCGTGGTGACAGCCCGCGAGGCCGTGAAGTGCGGCAAGTGTGTCGTCATCGGCCTGCAGAGCACCGGGGAGGCGCGCACCCTCGACCAGCTCGAGAGGGACGACGGGGACCTCACAGACTTCGTGTCCACCGCCAAGGGGGTGCTGCAGACGCTTGTGGAGAAGCACTTCCCGGCGCCGGACCGCGACCGCATCAACCGGCTGCTCGGGCTCGGCGCCAACAGGCCGCCGCCCCCCGCGCACGCCGCGCTCCCCGCGCCCCCGCACAACGGCACCAACGGTCGGGACGACGACGCCAACACGTCCAAGAGAAAGTTGACGTCACGCCAGCAAGCGAACGCCGCGGCCAAGCGCGCCCGCGGGGGCTCCTCGTCCGACGAGTTCGTCCGCGGCGGCTCCTCCGACGAGCTGGAGATGGAGTCGGACGAGGAGCAGCAGCGGGCCGCCTCCGACTCCGACCTCAGCGACTTCAACCCCTTCAGGGGCGGCAGCGACAGCGACGACGAACCCTGGCTGAATCGCAAAAAGAAACCAAAGAAGAAGAAGGCTGCATCTCCCAAGAAGAAGACCTCGACCACGCAGGACAAGATCGAGAACATGTTCACCCGCAAGAACAGCGCGCCCCCCGCCCCCACCCTCACCGTCACCACCAGCTCCGGCGCCAGCCTCACCGGGACGCCGGTCGGCACCAACGGCCTGTATCTAGGTCGCAGCCGCAGCAGCGCGCCGCCCCGGTCGGCCATCGAGCGCGCGTGCAGCATGAAGGAGGAGCTGCTGGCGGCCGTGCACCGGCTGGGTCGCCGCCTGCCGCCCAACACGCTCGACCAGCTCGTCGACGAGCTGGGGGGCACCGACAACGTGGCTGAGATGACAGGTCGCAAAGGGCGCGTGGTCCAAACAGAGGACGGACAGATCTTATACGAGAGCCGATCCGAAGCGGACGTGCCGCTCGAGACCCTGAACCTGACTGAGAAGCAAAGGTTCATGGATGGCGAGAAGGACGTGGCCATCATCTCTGAGGCCGCCTCCAGTGGTATCTCATTACAGAGTGATCGGAGGGCGAGGAATCAGAGGCGACGAGTTCACATCACACTAGAGCTGCCGTGGTCCGCGGACAGAGCCATACAACAGTTCGGGCGGACCCACCGCTCCAACCAGGTGAACGCCCCGGAGTACATATTCCTGATCTCGGACCTGGCCGGGGAGAGGCGGTTCGCGTGCACCGTCGCCAAGCGCCTGGAGTCGCTGGGTGCCCTCACGCACGGGGACCGCAGGGCCACCGAGACCAGGGACCTGTCGCAGTTCAACATCGACAACAAGTATGGTCGCACGGCGCTGGAGGCGGTGATGCGAGCCATCATGAAGTACGAGACGCCGCTGGTGCCCGCGCCCGCCGACTACCACGGAGACTTCTTCCAGGACGTGGCCAGCGCGCTCGTCGGCGTGGGGCTCATCGTCAACAGCGACGCCGCGCCCGGCGTGCTCGCGCTCGACAAGGACTATAACAACATGTCGAAGTTCTTGAACAGGATATTGGGCATGCCGGTGGAACTGCAGAACAGACTCTTCAAATACTTCACGGACACGCTCGCCGCCGTTATGGAACAAGCGAAGAGGAGCGGTCGGTTCGACCTGGGCATCCTGGACCTGGGCAGCGCGGGCGAGACCGTGCGGCGCCTGCGCTGCGTGCGCTTCCTGCGCCGCCACGCCACCGGACGCGCGCCCGTCGAGCTGCACACCGTCTCCTCCGAGCGCGGCATGGAATGGACTGAGGCTCTGGAGAAGCACTCGGAGCTGACGGGCCCCAAGGAGGGCTTCTACCTGTCGCAGCAGGCCCGCAACAGCAAGCACACCGCCGTGCTCTGCGCCGCCGCGCTCACCAACTCCAAGAAGGACAAGCTCAGCAAGAAGGACCTCATGTTTCAGATATACAGGCCGAACACTGGTCTGCAGCTGCGGCTGGAGTCGCTGGCGGAGATCGAGAAGAAGTACCGCAAGGTGGATGCGGACGAGGCGGAGAGCTGGTGGCGCGCGCAGCACGCGGCGTCGCTGCGGGTGTGCTCGCACGCGTACTGGCGGGACGCGTGCCGCGCCGCCGACTGCGAGGTGGGGCTGCGGCGCCGCCTGCACCACGTGCTGGCCGGCTCCGTGCTGGCCGTGTGGGCGCGCGTGGAGGCCACGCTGGCCGCGCGCTCGCAGCTGCACAAGATGCAGGTCGTGCGCATCAAGACCGACGACGGACTCAAGATTGTCGGCACGCTGATCCCCAAGAACTGCGTTGAGCCGCTGAAGGAGTCCCTCGCCGCGGACGCCGTCTCCGTCACGGAGCAGACCTTCGAGCAGACGGAGCTACACTGA
Protein
MSRRAYAEDPAPSDDSDFDDEEDPDNLEVPGGGKTLAAAARMADKKVPQATAPQVTVRPTTNPLAAPSGVAFGQTHNGKPIKIQASSLTKPFNLLNLPGVGRGGMGAAMAPFAAGSSAAFNAASLLAQMEMVGITGVNSYLLQNLNQIIAAGMGGSMLGQFSQQLPPWAGQIKPGMWHQDKMPGEEEEVDDEEMGVAETYADYMPPKVKLGRKHPDPVVETASLSSVEPVDVTYKLSLPDETIRGGLLSALQLEAVVYASQAHEHTLPDGTRAGFLIGDGAGVGKGRTIAGIIFENYIKGRKRAIWVSVSNDLKYDAERDLRDIGAGKIEVYPLNKFKYAKISSAVNGNVKKGVVFATYSSLIGETASNNTKYRSRLKQLLQWCGEDFDGVIVFDECHKAKNLCPVGSGKATKTGLTALELQNKLPKARVVYASATGASEPRNMAYMVRLGIWGEGTPFPTFMDFINAVEKRGVGAMEIVAMDMKLRGMYIARQLSFHGVSFRIEEVPLSDAFKETYDKSVALWVEAMQRFTEAAELIDAESRMRKTMWGQFWSAHQRFFKYLCIAAKVNHAVVTAREAVKCGKCVVIGLQSTGEARTLDQLERDDGDLTDFVSTAKGVLQTLVEKHFPAPDRDRINRLLGLGANRPPPPAHAALPAPPHNGTNGRDDDANTSKRKLTSRQQANAAAKRARGGSSSDEFVRGGSSDELEMESDEEQQRAASDSDLSDFNPFRGGSDSDDEPWLNRKKKPKKKKAASPKKKTSTTQDKIENMFTRKNSAPPAPTLTVTTSSGASLTGTPVGTNGLYLGRSRSSAPPRSAIERACSMKEELLAAVHRLGRRLPPNTLDQLVDELGGTDNVAEMTGRKGRVVQTEDGQILYESRSEADVPLETLNLTEKQRFMDGEKDVAIISEAASSGISLQSDRRARNQRRRVHITLELPWSADRAIQQFGRTHRSNQVNAPEYIFLISDLAGERRFACTVAKRLESLGALTHGDRRATETRDLSQFNIDNKYGRTALEAVMRAIMKYETPLVPAPADYHGDFFQDVASALVGVGLIVNSDAAPGVLALDKDYNNMSKFLNRILGMPVELQNRLFKYFTDTLAAVMEQAKRSGRFDLGILDLGSAGETVRRLRCVRFLRRHATGRAPVELHTVSSERGMEWTEALEKHSELTGPKEGFYLSQQARNSKHTAVLCAAALTNSKKDKLSKKDLMFQIYRPNTGLQLRLESLAEIEKKYRKVDADEAESWWRAQHAASLRVCSHAYWRDACRAADCEVGLRRRLHHVLAGSVLAVWARVEATLAARSQLHKMQVVRIKTDDGLKIVGTLIPKNCVEPLKESLAADAVSVTEQTFEQTELH
Summary
Uniprot
H9JU67
A0A2H1V3M8
A0A437BF95
A0A194R707
A0A194Q0A0
A0A212FN27
+ More
A0A139WGA0 A0A139WGU1 A0A139WGK9 A0A2A4K1W6 A0A026VXX2 A0A195DCR5 A0A195FU75 A0A195BCH9 E2AF27 E2BZX5 A0A1Q3F8P7 A0A195C684 A0A1Q3F8H9 W4VRF7 A0A2J7R848 A0A067R5Q0 A0A1B6K2L0 A0A1S4G7A3 A0A1B6E4P7 A0A1B0G3Z8 Q17PW5 A0A1A9W185 A0A0K8VQV2 A0A0K8VCA4 Q7QHI7 A0A1I8M306 W5J8F3 A0A2M4B9D3 A0A182FVI1 A0A2M4B9X2 A0A0A9WXT7 A0A182Q7B7 A0A0A9Y5M8 A0A182GST2 W8AKS6 W8AWV2 W8B7S2 A0A034V3Q7 A0A182HDV4 W8BHU6 A0A1A9VB05 A0A1B0A9Z8 A0A1J1HG53 A0A1J1HKH9 T1HUJ5 A0A1A9XC27 A0A1B0BGJ3 A0A2S2PFJ0 A0A2M3Z5F2 L7M738 A0A131XCF2 A0A224YD96 A0A131YWY5 A0A2R5LAR5 E0VMM9 A0A146LWG7 A0A0K8WAS8 A0A087UXI6 A0A1E1XBV7 A0A1E1XQW7 A0A2P6LI19 V9K9N7 A0A401SZB6 A0A091TBB9 H9GMG0 A0A1L8HQM1 A0A2Y9RSW4 W5MEV6 F6YBA0 A0A0P7U5I5 A0A1W5A6Y6 A0A1W5A696 A0A0K8S4W6 A0A0T6B7G6
A0A139WGA0 A0A139WGU1 A0A139WGK9 A0A2A4K1W6 A0A026VXX2 A0A195DCR5 A0A195FU75 A0A195BCH9 E2AF27 E2BZX5 A0A1Q3F8P7 A0A195C684 A0A1Q3F8H9 W4VRF7 A0A2J7R848 A0A067R5Q0 A0A1B6K2L0 A0A1S4G7A3 A0A1B6E4P7 A0A1B0G3Z8 Q17PW5 A0A1A9W185 A0A0K8VQV2 A0A0K8VCA4 Q7QHI7 A0A1I8M306 W5J8F3 A0A2M4B9D3 A0A182FVI1 A0A2M4B9X2 A0A0A9WXT7 A0A182Q7B7 A0A0A9Y5M8 A0A182GST2 W8AKS6 W8AWV2 W8B7S2 A0A034V3Q7 A0A182HDV4 W8BHU6 A0A1A9VB05 A0A1B0A9Z8 A0A1J1HG53 A0A1J1HKH9 T1HUJ5 A0A1A9XC27 A0A1B0BGJ3 A0A2S2PFJ0 A0A2M3Z5F2 L7M738 A0A131XCF2 A0A224YD96 A0A131YWY5 A0A2R5LAR5 E0VMM9 A0A146LWG7 A0A0K8WAS8 A0A087UXI6 A0A1E1XBV7 A0A1E1XQW7 A0A2P6LI19 V9K9N7 A0A401SZB6 A0A091TBB9 H9GMG0 A0A1L8HQM1 A0A2Y9RSW4 W5MEV6 F6YBA0 A0A0P7U5I5 A0A1W5A6Y6 A0A1W5A696 A0A0K8S4W6 A0A0T6B7G6
Pubmed
EMBL
BABH01004162
BABH01004163
BABH01004164
BABH01004165
BABH01004166
BABH01004167
+ More
BABH01004168 BABH01004169 ODYU01000525 SOQ35437.1 RSAL01000071 RVE49107.1 KQ460878 KPJ11651.1 KQ459582 KPI99007.1 AGBW02007651 OWR55109.1 KQ971345 KYB27028.1 KYB27027.1 KYB27029.1 NWSH01000261 PCG77918.1 KK107796 QOIP01000013 EZA47704.1 RLU15499.1 KQ980989 KYN10676.1 KQ981264 KYN43976.1 KQ976528 KYM81900.1 GL438984 EFN67982.1 GL451708 EFN78752.1 GFDL01011148 JAV23897.1 KQ978220 KYM96374.1 GFDL01011170 JAV23875.1 GANO01003817 JAB56054.1 NEVH01006723 PNF37003.1 KK852685 KDR18510.1 GECU01002323 JAT05384.1 GEDC01004414 JAS32884.1 CCAG010005646 CCAG010005647 CH477188 EAT48829.1 GDHF01011062 JAI41252.1 GDHF01015842 JAI36472.1 AAAB01008816 EAA05169.4 ADMH02002067 ETN59668.1 GGFJ01000495 MBW49636.1 GGFJ01000650 MBW49791.1 GBHO01033974 JAG09630.1 AXCN02001714 GBHO01017201 JAG26403.1 JXUM01085204 JXUM01085205 JXUM01085206 KQ563487 KXJ73756.1 GAMC01017280 JAB89275.1 GAMC01017282 JAB89273.1 GAMC01017279 JAB89276.1 GAKP01021026 JAC37926.1 JXUM01130476 JXUM01130477 KQ567584 KXJ69359.1 GAMC01017281 JAB89274.1 CVRI01000002 CRK86967.1 CRK86966.1 ACPB03011481 JXJN01013884 GGMR01015359 MBY27978.1 GGFM01002998 MBW23749.1 GACK01005362 JAA59672.1 GEFH01003972 JAP64609.1 GFPF01004532 MAA15678.1 GEDV01004848 JAP83709.1 GGLE01002453 MBY06579.1 DS235317 EEB14635.1 GDHC01007939 JAQ10690.1 GDHF01004078 JAI48236.1 KK122158 KFM82075.1 GFAC01002587 JAT96601.1 GFAA01001704 JAU01731.1 MWRG01000049 PRD38236.1 JW861819 AFO94336.1 BEZZ01000738 GCC35731.1 KK444120 KFQ71127.1 CM004467 OCT98378.1 AHAT01005140 AAMC01042902 AAMC01042903 AAMC01042904 JARO02008539 KPP62592.1 GBRD01017539 JAG48288.1 LJIG01009333 KRT83290.1
BABH01004168 BABH01004169 ODYU01000525 SOQ35437.1 RSAL01000071 RVE49107.1 KQ460878 KPJ11651.1 KQ459582 KPI99007.1 AGBW02007651 OWR55109.1 KQ971345 KYB27028.1 KYB27027.1 KYB27029.1 NWSH01000261 PCG77918.1 KK107796 QOIP01000013 EZA47704.1 RLU15499.1 KQ980989 KYN10676.1 KQ981264 KYN43976.1 KQ976528 KYM81900.1 GL438984 EFN67982.1 GL451708 EFN78752.1 GFDL01011148 JAV23897.1 KQ978220 KYM96374.1 GFDL01011170 JAV23875.1 GANO01003817 JAB56054.1 NEVH01006723 PNF37003.1 KK852685 KDR18510.1 GECU01002323 JAT05384.1 GEDC01004414 JAS32884.1 CCAG010005646 CCAG010005647 CH477188 EAT48829.1 GDHF01011062 JAI41252.1 GDHF01015842 JAI36472.1 AAAB01008816 EAA05169.4 ADMH02002067 ETN59668.1 GGFJ01000495 MBW49636.1 GGFJ01000650 MBW49791.1 GBHO01033974 JAG09630.1 AXCN02001714 GBHO01017201 JAG26403.1 JXUM01085204 JXUM01085205 JXUM01085206 KQ563487 KXJ73756.1 GAMC01017280 JAB89275.1 GAMC01017282 JAB89273.1 GAMC01017279 JAB89276.1 GAKP01021026 JAC37926.1 JXUM01130476 JXUM01130477 KQ567584 KXJ69359.1 GAMC01017281 JAB89274.1 CVRI01000002 CRK86967.1 CRK86966.1 ACPB03011481 JXJN01013884 GGMR01015359 MBY27978.1 GGFM01002998 MBW23749.1 GACK01005362 JAA59672.1 GEFH01003972 JAP64609.1 GFPF01004532 MAA15678.1 GEDV01004848 JAP83709.1 GGLE01002453 MBY06579.1 DS235317 EEB14635.1 GDHC01007939 JAQ10690.1 GDHF01004078 JAI48236.1 KK122158 KFM82075.1 GFAC01002587 JAT96601.1 GFAA01001704 JAU01731.1 MWRG01000049 PRD38236.1 JW861819 AFO94336.1 BEZZ01000738 GCC35731.1 KK444120 KFQ71127.1 CM004467 OCT98378.1 AHAT01005140 AAMC01042902 AAMC01042903 AAMC01042904 JARO02008539 KPP62592.1 GBRD01017539 JAG48288.1 LJIG01009333 KRT83290.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000053268
UP000007151
UP000007266
+ More
UP000218220 UP000053097 UP000279307 UP000078492 UP000078541 UP000078540 UP000000311 UP000008237 UP000078542 UP000235965 UP000027135 UP000092444 UP000008820 UP000091820 UP000007062 UP000095301 UP000000673 UP000069272 UP000075886 UP000069940 UP000249989 UP000078200 UP000092445 UP000183832 UP000015103 UP000092443 UP000092460 UP000009046 UP000054359 UP000287033 UP000001646 UP000186698 UP000248480 UP000018468 UP000008143 UP000034805 UP000192224
UP000218220 UP000053097 UP000279307 UP000078492 UP000078541 UP000078540 UP000000311 UP000008237 UP000078542 UP000235965 UP000027135 UP000092444 UP000008820 UP000091820 UP000007062 UP000095301 UP000000673 UP000069272 UP000075886 UP000069940 UP000249989 UP000078200 UP000092445 UP000183832 UP000015103 UP000092443 UP000092460 UP000009046 UP000054359 UP000287033 UP000001646 UP000186698 UP000248480 UP000018468 UP000008143 UP000034805 UP000192224
Interpro
Gene 3D
ProteinModelPortal
H9JU67
A0A2H1V3M8
A0A437BF95
A0A194R707
A0A194Q0A0
A0A212FN27
+ More
A0A139WGA0 A0A139WGU1 A0A139WGK9 A0A2A4K1W6 A0A026VXX2 A0A195DCR5 A0A195FU75 A0A195BCH9 E2AF27 E2BZX5 A0A1Q3F8P7 A0A195C684 A0A1Q3F8H9 W4VRF7 A0A2J7R848 A0A067R5Q0 A0A1B6K2L0 A0A1S4G7A3 A0A1B6E4P7 A0A1B0G3Z8 Q17PW5 A0A1A9W185 A0A0K8VQV2 A0A0K8VCA4 Q7QHI7 A0A1I8M306 W5J8F3 A0A2M4B9D3 A0A182FVI1 A0A2M4B9X2 A0A0A9WXT7 A0A182Q7B7 A0A0A9Y5M8 A0A182GST2 W8AKS6 W8AWV2 W8B7S2 A0A034V3Q7 A0A182HDV4 W8BHU6 A0A1A9VB05 A0A1B0A9Z8 A0A1J1HG53 A0A1J1HKH9 T1HUJ5 A0A1A9XC27 A0A1B0BGJ3 A0A2S2PFJ0 A0A2M3Z5F2 L7M738 A0A131XCF2 A0A224YD96 A0A131YWY5 A0A2R5LAR5 E0VMM9 A0A146LWG7 A0A0K8WAS8 A0A087UXI6 A0A1E1XBV7 A0A1E1XQW7 A0A2P6LI19 V9K9N7 A0A401SZB6 A0A091TBB9 H9GMG0 A0A1L8HQM1 A0A2Y9RSW4 W5MEV6 F6YBA0 A0A0P7U5I5 A0A1W5A6Y6 A0A1W5A696 A0A0K8S4W6 A0A0T6B7G6
A0A139WGA0 A0A139WGU1 A0A139WGK9 A0A2A4K1W6 A0A026VXX2 A0A195DCR5 A0A195FU75 A0A195BCH9 E2AF27 E2BZX5 A0A1Q3F8P7 A0A195C684 A0A1Q3F8H9 W4VRF7 A0A2J7R848 A0A067R5Q0 A0A1B6K2L0 A0A1S4G7A3 A0A1B6E4P7 A0A1B0G3Z8 Q17PW5 A0A1A9W185 A0A0K8VQV2 A0A0K8VCA4 Q7QHI7 A0A1I8M306 W5J8F3 A0A2M4B9D3 A0A182FVI1 A0A2M4B9X2 A0A0A9WXT7 A0A182Q7B7 A0A0A9Y5M8 A0A182GST2 W8AKS6 W8AWV2 W8B7S2 A0A034V3Q7 A0A182HDV4 W8BHU6 A0A1A9VB05 A0A1B0A9Z8 A0A1J1HG53 A0A1J1HKH9 T1HUJ5 A0A1A9XC27 A0A1B0BGJ3 A0A2S2PFJ0 A0A2M3Z5F2 L7M738 A0A131XCF2 A0A224YD96 A0A131YWY5 A0A2R5LAR5 E0VMM9 A0A146LWG7 A0A0K8WAS8 A0A087UXI6 A0A1E1XBV7 A0A1E1XQW7 A0A2P6LI19 V9K9N7 A0A401SZB6 A0A091TBB9 H9GMG0 A0A1L8HQM1 A0A2Y9RSW4 W5MEV6 F6YBA0 A0A0P7U5I5 A0A1W5A6Y6 A0A1W5A696 A0A0K8S4W6 A0A0T6B7G6
Ontologies
PANTHER
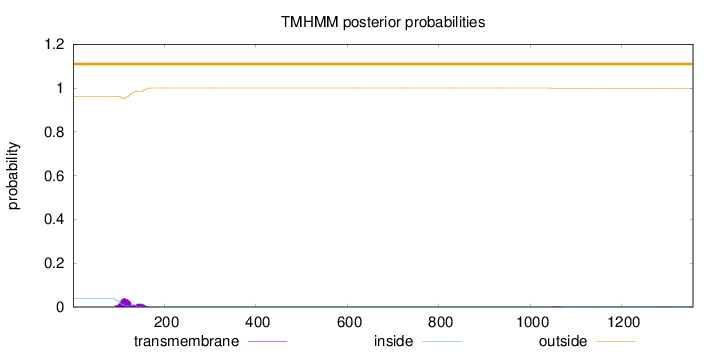
Topology
Length:
1356
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.37522
Exp number, first 60 AAs:
0.00014
Total prob of N-in:
0.03810
outside
1 - 1356
Population Genetic Test Statistics
Pi
223.314829
Theta
162.470243
Tajima's D
1.261579
CLR
0.322039
CSRT
0.730013499325034
Interpretation
Uncertain
