Gene
KWMTBOMO09425
Pre Gene Modal
BGIBMGA012775
Annotation
PREDICTED:_probable_ATP-dependent_RNA_helicase_DDX55_homolog_[Papilio_polytes]
Full name
RNA helicase
+ More
Probable ATP-dependent RNA helicase DDX55 homolog
Probable ATP-dependent RNA helicase DDX55 homolog
Location in the cell
Cytoplasmic Reliability : 1.478 Mitochondrial Reliability : 1.144 Nuclear Reliability : 1.155
Sequence
CDS
ATGGCCAACAAAGATTGGTCTAAAGTTCAGCCCCCATTATCCAAGCCAGTGTTGAACTGTATTAATAAACATGGTTTTAAAACTATGACTCCGATTCAAGCAGCCGTAATTCCTCTATTATTGTCCTGCAAAGATGTTGTGGCTGAAGCGGTGACTGGATCGGGAAAAACTCTCGCATTTGTCGTCCCGATGCTGGAAATGCTCTTGAAAAAGAAGAAAGATGGACCTTTGAGAAAGGATTTCGTTTACGTAGTCATAATTTCGCCTACAAGGGAACTGGCTACGCAAATATTTAAAGTAATAGAAGATTTCTTACAAGAACCAGAATTAGCCAGTACTACCTTGGCCCTATTAGTCGGTGGTCGTCCCATAGAGGTTGATGCGCTTGCCTTACAGAAAGGAGCTGATATAGCTGTATGCACTCCGGGTAGACTAGAAGACTTGTTGTCTGAAAGGAAACAGTTGAATCTTGCCGGAAGACTGAAAGAGTTGGAACTCTTAGTTCTGGATGAAGCGGACCGTCTACTAGACCTTGGCTTCTCGGGCACACTCACAACCATCCTACAATATTTACCGAGACAACGTCGTACTGGTCTCTTTTCGGCAACACAAACCAAAGAACTGCAGGACTTAGTAAGAGCTGGTCTTCGGAATCCTGTTGTTGTGAGCGTGAAAGAAAAGTCTACAATCAGTACTCCCTTGCTGTTGGAGACTTATTACGTAATTGTTGAAGCTCAGGATAAATTTTTGTTCCTTTTGAACTTTATCAGAAATAGAAAGATAGCTAAAGGTTTATTTTTTCTACCAACGTGTGCATGTGTGGATTACTGGACGGATGTGCTACCCGTATTCCTGCCTGACATTAAAATATTTGGATTACACGGCAAAATGAAGCAGAAACGACACAAAATATTAGAGAAGTTCCGGCAGGCTGATAACACTATCTTGCTTTGTACAGATTTGTTGGCCAGGGGCCTGGACATTCCAGAGGTGGAGTGGGTGTTGCAGTGGGAGCCCCCATGCCAACCCTCCGCTCTCGTGCACCGCGCGGGCAGAGCAGCGAGGGGCGGAGCCCGGGGCTGCGCCCTGCTACCGCTGCTCACCACAGAGGACGCTTACGTGCCCTTCATAAAGACTAATCAAATGGTCGAGTTGAAAGACTGGAGGGAATCTGACGATCAAATCAAAATTACGGACAAGTTAAGGGAAAGGGTGCTCGGCATCCTCCACGAGCAGCAGAAGAAAGACCGCGCCATTTTCGACAAAGGGCAGCGAGCGTTCGTCTCCCACATGCGAGCCTACACCAAACACGAATGCAACCTGCTGCTCCAGTTCAAGGAGCTTCCTCTGGGGCACATCGCCACCAGCTACGGGCTCCTCAAACTGCCTCTCATGCCAGAGATCAAACCGGAGCACAAGCAGCAGTTTGTCGGCCCCAAAGAGGAGATAGACTTTAACACAATTCAGTACAGGGATAAGCAGAAGGAGACGAGTCGCCTGCAGAAGTTGGAGGAGTATAAGAAGAGCGGGGTTTGGCCGTCGAAGAAAAAAAAGAAAATGGTCCAAACACTCCCGTGGGAACAAGCGAAACAGAACAAAGAAGAAAGGAAAGAGAAAAGGAAGAAACGTAAAGAGTTGAAACAGAAGAATCCAGAACTTAAGAAAGGAAAGAAGAGGAAAGTGATAACGCAAGAGGAACTGGACGCGCTGGCCGCCGACGTCGCGCTCATAAAGAAGTTAAAGAAACGTAAAATAACACAGGATCAGTTCGATAGAGAGTTTGAAGCTGATTAA
Protein
MANKDWSKVQPPLSKPVLNCINKHGFKTMTPIQAAVIPLLLSCKDVVAEAVTGSGKTLAFVVPMLEMLLKKKKDGPLRKDFVYVVIISPTRELATQIFKVIEDFLQEPELASTTLALLVGGRPIEVDALALQKGADIAVCTPGRLEDLLSERKQLNLAGRLKELELLVLDEADRLLDLGFSGTLTTILQYLPRQRRTGLFSATQTKELQDLVRAGLRNPVVVSVKEKSTISTPLLLETYYVIVEAQDKFLFLLNFIRNRKIAKGLFFLPTCACVDYWTDVLPVFLPDIKIFGLHGKMKQKRHKILEKFRQADNTILLCTDLLARGLDIPEVEWVLQWEPPCQPSALVHRAGRAARGGARGCALLPLLTTEDAYVPFIKTNQMVELKDWRESDDQIKITDKLRERVLGILHEQQKKDRAIFDKGQRAFVSHMRAYTKHECNLLLQFKELPLGHIATSYGLLKLPLMPEIKPEHKQQFVGPKEEIDFNTIQYRDKQKETSRLQKLEEYKKSGVWPSKKKKKMVQTLPWEQAKQNKEERKEKRKKRKELKQKNPELKKGKKRKVITQEELDALAADVALIKKLKKRKITQDQFDREFEAD
Summary
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Similarity
Belongs to the DEAD box helicase family.
Belongs to the DEAD box helicase family. DDX55/SPB4 subfamily.
Belongs to the DEAD box helicase family. DDX55/SPB4 subfamily.
Keywords
ATP-binding
Coiled coil
Complete proteome
Helicase
Hydrolase
Nucleotide-binding
Reference proteome
RNA-binding
Feature
chain Probable ATP-dependent RNA helicase DDX55 homolog
Uniprot
H9JTB3
A0A2H1VT91
A0A194Q699
A0A194RS99
A0A437BFE2
A0A212FMD0
+ More
A0A182VW07 A0A182MNG1 W5J575 A0A182GVI8 A0A2M4BHL3 A0A2M4ANF2 A0A2M4BI86 A0A182GXM1 A0A2M4BHJ5 A0A182FS29 Q175I3 A0A336MHP8 B0WK71 A0A182QRE8 A0A182YHX4 A0A182K253 A0A1Q3EXT9 A0A182PRV3 A0A182L8K5 A0A182NQS2 A0A182HKB6 A0A182X9D0 A0A0M4EN68 Q7PX55 A0A182THT4 A0A182V0I3 A0A182RSD3 A0A1B0BKJ9 A0A1B0A549 A0A0L0C3X8 A0A1B0A5N3 A0A1A9Y6F5 A0A1A9UNF7 A0A182JDX9 A0A1B0FRH1 W8B1K3 A0A1I8NH73 A0A1A9WGS6 B4KA13 A0A084W028 A0A1Q3EXS9 A0A2M4AMY6 A0A0K8WFU2 A0A2M4APF4 A0A1I8QD21 A0A1J1I7W0 B4LVT6 B4NAK4 A0A1W4VH96 B4HLN4 A0A3B0KCS2 B3LYX3 Q298Q2 B4G4F7 B4QYN2 Q9VHU1 B4PT28 B3NZV5 A0A0M8ZRM3 E2C3Z6 E2AAH9 A0A088AUQ5 B4JXW1 A0A154PM57 A0A1W4W348 A0A151WUW6 A0A2A3ET37 A0A195CS30 D6WYU0 A0A158NW40 E9JAY7 F4X2F7 N6SVI4 U4U740 A0A1Y1LF35 A0A232EV27 K7IS49 A0A1B6JTZ4 A0A067R2Z7 A0A1B6GFY9 A0A2S2Q6H4 J9K0V4 A0A2J7QH41 A0A2P8YC27 E0VGY5 A0A2H8TN29 A7S4S3 A0A0A9VVU9 A0A1B6CXC2 R4WJS8 C3YD28 A0A2R7X3F6 A0A0P4W7H0 A0A026VVP3
A0A182VW07 A0A182MNG1 W5J575 A0A182GVI8 A0A2M4BHL3 A0A2M4ANF2 A0A2M4BI86 A0A182GXM1 A0A2M4BHJ5 A0A182FS29 Q175I3 A0A336MHP8 B0WK71 A0A182QRE8 A0A182YHX4 A0A182K253 A0A1Q3EXT9 A0A182PRV3 A0A182L8K5 A0A182NQS2 A0A182HKB6 A0A182X9D0 A0A0M4EN68 Q7PX55 A0A182THT4 A0A182V0I3 A0A182RSD3 A0A1B0BKJ9 A0A1B0A549 A0A0L0C3X8 A0A1B0A5N3 A0A1A9Y6F5 A0A1A9UNF7 A0A182JDX9 A0A1B0FRH1 W8B1K3 A0A1I8NH73 A0A1A9WGS6 B4KA13 A0A084W028 A0A1Q3EXS9 A0A2M4AMY6 A0A0K8WFU2 A0A2M4APF4 A0A1I8QD21 A0A1J1I7W0 B4LVT6 B4NAK4 A0A1W4VH96 B4HLN4 A0A3B0KCS2 B3LYX3 Q298Q2 B4G4F7 B4QYN2 Q9VHU1 B4PT28 B3NZV5 A0A0M8ZRM3 E2C3Z6 E2AAH9 A0A088AUQ5 B4JXW1 A0A154PM57 A0A1W4W348 A0A151WUW6 A0A2A3ET37 A0A195CS30 D6WYU0 A0A158NW40 E9JAY7 F4X2F7 N6SVI4 U4U740 A0A1Y1LF35 A0A232EV27 K7IS49 A0A1B6JTZ4 A0A067R2Z7 A0A1B6GFY9 A0A2S2Q6H4 J9K0V4 A0A2J7QH41 A0A2P8YC27 E0VGY5 A0A2H8TN29 A7S4S3 A0A0A9VVU9 A0A1B6CXC2 R4WJS8 C3YD28 A0A2R7X3F6 A0A0P4W7H0 A0A026VVP3
EC Number
3.6.4.13
Pubmed
19121390
26354079
22118469
20920257
23761445
26483478
+ More
17510324 25244985 20966253 12364791 14747013 17210077 26108605 24495485 25315136 17994087 24438588 15632085 10731132 12537572 12537569 17550304 20798317 18362917 19820115 21347285 21282665 21719571 23537049 28004739 28648823 20075255 24845553 29403074 20566863 17615350 25401762 26823975 23691247 18563158 24508170
17510324 25244985 20966253 12364791 14747013 17210077 26108605 24495485 25315136 17994087 24438588 15632085 10731132 12537572 12537569 17550304 20798317 18362917 19820115 21347285 21282665 21719571 23537049 28004739 28648823 20075255 24845553 29403074 20566863 17615350 25401762 26823975 23691247 18563158 24508170
EMBL
BABH01004175
ODYU01004311
SOQ44050.1
KQ459582
KPI98935.1
KQ459700
+ More
KPJ20352.1 RSAL01000071 RVE49101.1 AGBW02007653 OWR54886.1 AXCM01001239 ADMH02002101 ETN59131.1 JXUM01090920 KQ563875 KXJ73125.1 GGFJ01003386 MBW52527.1 GGFK01008988 MBW42309.1 GGFJ01003387 MBW52528.1 JXUM01095800 KQ564219 KXJ72571.1 GGFJ01003385 MBW52526.1 CH477400 EAT41737.1 UFQT01000590 SSX25548.1 DS231968 EDS29645.1 AXCN02001501 GFDL01014921 JAV20124.1 APCN01000825 CP012526 ALC47002.1 AAAB01008987 EAA01795.5 JXJN01015972 JRES01000936 KNC27058.1 CCAG010004927 GAMC01019434 JAB87121.1 CH933806 EDW16688.1 ATLV01019081 KE525262 KFB43572.1 GFDL01014931 JAV20114.1 GGFK01008823 MBW42144.1 GDHF01002357 JAI49957.1 GGFK01009167 MBW42488.1 CVRI01000039 CRK94489.1 CH940650 EDW67541.1 CH964232 EDW80818.1 CH480815 EDW43061.1 OUUW01000007 SPP82811.1 CH902617 EDV44089.1 CM000070 EAL27903.2 CH479179 EDW24505.1 CM000364 EDX13790.1 AE014297 AY051513 CM000160 EDW96489.1 CH954181 EDV49953.1 KQ435883 KOX69710.1 GL452364 EFN77382.1 GL438125 EFN69580.1 CH916377 EDV90523.1 KQ434954 KZC12544.1 KQ982721 KYQ51653.1 KZ288192 PBC34211.1 KQ977329 KYN03543.1 KQ971357 EFA08391.1 ADTU01027781 ADTU01027782 GL770679 EFZ10065.1 GL888578 EGI59382.1 APGK01054966 APGK01054970 KB741256 ENN71750.1 ENN71754.1 KB631749 ERL85775.1 GEZM01057600 JAV72233.1 NNAY01002058 OXU22212.1 GECU01005035 JAT02672.1 KK853042 KDR12156.1 GECZ01008421 JAS61348.1 GGMS01004126 MBY73329.1 ABLF02036425 NEVH01014358 PNF27905.1 PYGN01000713 PSN41815.1 DS235154 EEB12641.1 GFXV01003624 MBW15429.1 DS469579 EDO41335.1 GBHO01043242 GBRD01003086 GDHC01016998 JAG00362.1 JAG62735.1 JAQ01631.1 GEDC01019425 JAS17873.1 AK417855 BAN21070.1 GG666503 EEN61639.1 KK856724 PTY26328.1 GDRN01085388 JAI61342.1 KK107801 EZA47581.1
KPJ20352.1 RSAL01000071 RVE49101.1 AGBW02007653 OWR54886.1 AXCM01001239 ADMH02002101 ETN59131.1 JXUM01090920 KQ563875 KXJ73125.1 GGFJ01003386 MBW52527.1 GGFK01008988 MBW42309.1 GGFJ01003387 MBW52528.1 JXUM01095800 KQ564219 KXJ72571.1 GGFJ01003385 MBW52526.1 CH477400 EAT41737.1 UFQT01000590 SSX25548.1 DS231968 EDS29645.1 AXCN02001501 GFDL01014921 JAV20124.1 APCN01000825 CP012526 ALC47002.1 AAAB01008987 EAA01795.5 JXJN01015972 JRES01000936 KNC27058.1 CCAG010004927 GAMC01019434 JAB87121.1 CH933806 EDW16688.1 ATLV01019081 KE525262 KFB43572.1 GFDL01014931 JAV20114.1 GGFK01008823 MBW42144.1 GDHF01002357 JAI49957.1 GGFK01009167 MBW42488.1 CVRI01000039 CRK94489.1 CH940650 EDW67541.1 CH964232 EDW80818.1 CH480815 EDW43061.1 OUUW01000007 SPP82811.1 CH902617 EDV44089.1 CM000070 EAL27903.2 CH479179 EDW24505.1 CM000364 EDX13790.1 AE014297 AY051513 CM000160 EDW96489.1 CH954181 EDV49953.1 KQ435883 KOX69710.1 GL452364 EFN77382.1 GL438125 EFN69580.1 CH916377 EDV90523.1 KQ434954 KZC12544.1 KQ982721 KYQ51653.1 KZ288192 PBC34211.1 KQ977329 KYN03543.1 KQ971357 EFA08391.1 ADTU01027781 ADTU01027782 GL770679 EFZ10065.1 GL888578 EGI59382.1 APGK01054966 APGK01054970 KB741256 ENN71750.1 ENN71754.1 KB631749 ERL85775.1 GEZM01057600 JAV72233.1 NNAY01002058 OXU22212.1 GECU01005035 JAT02672.1 KK853042 KDR12156.1 GECZ01008421 JAS61348.1 GGMS01004126 MBY73329.1 ABLF02036425 NEVH01014358 PNF27905.1 PYGN01000713 PSN41815.1 DS235154 EEB12641.1 GFXV01003624 MBW15429.1 DS469579 EDO41335.1 GBHO01043242 GBRD01003086 GDHC01016998 JAG00362.1 JAG62735.1 JAQ01631.1 GEDC01019425 JAS17873.1 AK417855 BAN21070.1 GG666503 EEN61639.1 KK856724 PTY26328.1 GDRN01085388 JAI61342.1 KK107801 EZA47581.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000283053
UP000007151
UP000075920
+ More
UP000075883 UP000000673 UP000069940 UP000249989 UP000069272 UP000008820 UP000002320 UP000075886 UP000076408 UP000075881 UP000075885 UP000075882 UP000075884 UP000075840 UP000076407 UP000092553 UP000007062 UP000075902 UP000075903 UP000075900 UP000092460 UP000092445 UP000037069 UP000092443 UP000078200 UP000075880 UP000092444 UP000095301 UP000091820 UP000009192 UP000030765 UP000095300 UP000183832 UP000008792 UP000007798 UP000192221 UP000001292 UP000268350 UP000007801 UP000001819 UP000008744 UP000000304 UP000000803 UP000002282 UP000008711 UP000053105 UP000008237 UP000000311 UP000005203 UP000001070 UP000076502 UP000192223 UP000075809 UP000242457 UP000078542 UP000007266 UP000005205 UP000007755 UP000019118 UP000030742 UP000215335 UP000002358 UP000027135 UP000007819 UP000235965 UP000245037 UP000009046 UP000001593 UP000001554 UP000053097
UP000075883 UP000000673 UP000069940 UP000249989 UP000069272 UP000008820 UP000002320 UP000075886 UP000076408 UP000075881 UP000075885 UP000075882 UP000075884 UP000075840 UP000076407 UP000092553 UP000007062 UP000075902 UP000075903 UP000075900 UP000092460 UP000092445 UP000037069 UP000092443 UP000078200 UP000075880 UP000092444 UP000095301 UP000091820 UP000009192 UP000030765 UP000095300 UP000183832 UP000008792 UP000007798 UP000192221 UP000001292 UP000268350 UP000007801 UP000001819 UP000008744 UP000000304 UP000000803 UP000002282 UP000008711 UP000053105 UP000008237 UP000000311 UP000005203 UP000001070 UP000076502 UP000192223 UP000075809 UP000242457 UP000078542 UP000007266 UP000005205 UP000007755 UP000019118 UP000030742 UP000215335 UP000002358 UP000027135 UP000007819 UP000235965 UP000245037 UP000009046 UP000001593 UP000001554 UP000053097
Interpro
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
H9JTB3
A0A2H1VT91
A0A194Q699
A0A194RS99
A0A437BFE2
A0A212FMD0
+ More
A0A182VW07 A0A182MNG1 W5J575 A0A182GVI8 A0A2M4BHL3 A0A2M4ANF2 A0A2M4BI86 A0A182GXM1 A0A2M4BHJ5 A0A182FS29 Q175I3 A0A336MHP8 B0WK71 A0A182QRE8 A0A182YHX4 A0A182K253 A0A1Q3EXT9 A0A182PRV3 A0A182L8K5 A0A182NQS2 A0A182HKB6 A0A182X9D0 A0A0M4EN68 Q7PX55 A0A182THT4 A0A182V0I3 A0A182RSD3 A0A1B0BKJ9 A0A1B0A549 A0A0L0C3X8 A0A1B0A5N3 A0A1A9Y6F5 A0A1A9UNF7 A0A182JDX9 A0A1B0FRH1 W8B1K3 A0A1I8NH73 A0A1A9WGS6 B4KA13 A0A084W028 A0A1Q3EXS9 A0A2M4AMY6 A0A0K8WFU2 A0A2M4APF4 A0A1I8QD21 A0A1J1I7W0 B4LVT6 B4NAK4 A0A1W4VH96 B4HLN4 A0A3B0KCS2 B3LYX3 Q298Q2 B4G4F7 B4QYN2 Q9VHU1 B4PT28 B3NZV5 A0A0M8ZRM3 E2C3Z6 E2AAH9 A0A088AUQ5 B4JXW1 A0A154PM57 A0A1W4W348 A0A151WUW6 A0A2A3ET37 A0A195CS30 D6WYU0 A0A158NW40 E9JAY7 F4X2F7 N6SVI4 U4U740 A0A1Y1LF35 A0A232EV27 K7IS49 A0A1B6JTZ4 A0A067R2Z7 A0A1B6GFY9 A0A2S2Q6H4 J9K0V4 A0A2J7QH41 A0A2P8YC27 E0VGY5 A0A2H8TN29 A7S4S3 A0A0A9VVU9 A0A1B6CXC2 R4WJS8 C3YD28 A0A2R7X3F6 A0A0P4W7H0 A0A026VVP3
A0A182VW07 A0A182MNG1 W5J575 A0A182GVI8 A0A2M4BHL3 A0A2M4ANF2 A0A2M4BI86 A0A182GXM1 A0A2M4BHJ5 A0A182FS29 Q175I3 A0A336MHP8 B0WK71 A0A182QRE8 A0A182YHX4 A0A182K253 A0A1Q3EXT9 A0A182PRV3 A0A182L8K5 A0A182NQS2 A0A182HKB6 A0A182X9D0 A0A0M4EN68 Q7PX55 A0A182THT4 A0A182V0I3 A0A182RSD3 A0A1B0BKJ9 A0A1B0A549 A0A0L0C3X8 A0A1B0A5N3 A0A1A9Y6F5 A0A1A9UNF7 A0A182JDX9 A0A1B0FRH1 W8B1K3 A0A1I8NH73 A0A1A9WGS6 B4KA13 A0A084W028 A0A1Q3EXS9 A0A2M4AMY6 A0A0K8WFU2 A0A2M4APF4 A0A1I8QD21 A0A1J1I7W0 B4LVT6 B4NAK4 A0A1W4VH96 B4HLN4 A0A3B0KCS2 B3LYX3 Q298Q2 B4G4F7 B4QYN2 Q9VHU1 B4PT28 B3NZV5 A0A0M8ZRM3 E2C3Z6 E2AAH9 A0A088AUQ5 B4JXW1 A0A154PM57 A0A1W4W348 A0A151WUW6 A0A2A3ET37 A0A195CS30 D6WYU0 A0A158NW40 E9JAY7 F4X2F7 N6SVI4 U4U740 A0A1Y1LF35 A0A232EV27 K7IS49 A0A1B6JTZ4 A0A067R2Z7 A0A1B6GFY9 A0A2S2Q6H4 J9K0V4 A0A2J7QH41 A0A2P8YC27 E0VGY5 A0A2H8TN29 A7S4S3 A0A0A9VVU9 A0A1B6CXC2 R4WJS8 C3YD28 A0A2R7X3F6 A0A0P4W7H0 A0A026VVP3
PDB
6EM5
E-value=1.42801e-66,
Score=644
Ontologies
GO
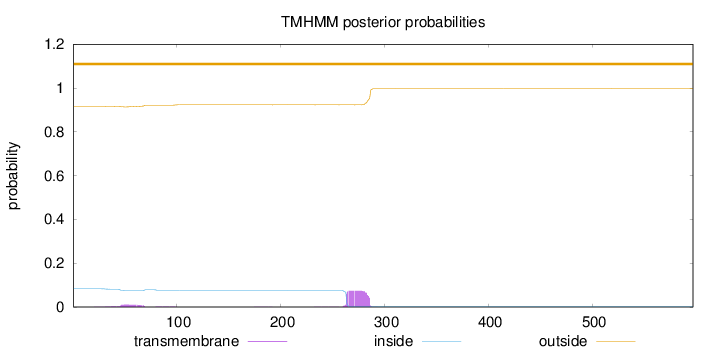
Topology
Length:
597
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.91857
Exp number, first 60 AAs:
0.18596
Total prob of N-in:
0.08336
outside
1 - 597
Population Genetic Test Statistics
Pi
237.552133
Theta
179.838633
Tajima's D
0.276176
CLR
143.20321
CSRT
0.454327283635818
Interpretation
Uncertain
