Gene
KWMTBOMO09424
Pre Gene Modal
BGIBMGA013082
Annotation
PREDICTED:_transmembrane_protein_134_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.215
Sequence
CDS
ATGACTAAACCGTTCGGAAACAGTGAGAAGCGTTTTTCGATCGACGATGCGTTCGAAGAAGAAACCGACGAAGCTATAAAAGTGTACGGCGCAACTGGAGACCGATCACCTTTGCAAAACAAATTCAAGAATGGTGGTGATTATATATCCAGCAAAACCTATAAATGCTCAGAAGATACAACCTCAAGGGATTCGGACTCCCTCATACATGAATATGTTGAGGCCACACAATCAATGTTCTGCTGGAACCATCCCAAAGTGAGAGAGAATTGGAAAACGGTATGTGCAGCTGTCATTTTGCTGATAGTTGGAGTTGGTCTACTTGGCATGGGAGCATTTGCGGTTGCTGAACCTGAAAACGGTCTCCAAGGTGCAGTGTTCTTCGTAGCGGGAATGATCTGCTTTGTGCCCGGTGCATATCATGTAGTCTATATCTGGCTAGCTGCCAGAGGACAGAGAGGTTACGATTTCTACCACCTTCCATTATTCACTTGA
Protein
MTKPFGNSEKRFSIDDAFEEETDEAIKVYGATGDRSPLQNKFKNGGDYISSKTYKCSEDTTSRDSDSLIHEYVEATQSMFCWNHPKVRENWKTVCAAVILLIVGVGLLGMGAFAVAEPENGLQGAVFFVAGMICFVPGAYHVVYIWLAARGQRGYDFYHLPLFT
Summary
Uniprot
H9JU70
A0A194Q049
A0A194RR46
A0A437BFF2
A0A0L7LMT7
A0A212FMD1
+ More
S4P753 A0A2A4K7L9 A0A1E1W935 A0A2H1VTB1 A0A2A4K777 A0A0K8TQ01 A0A182GQ91 W5JQC7 T1P8Q1 U5EQQ6 A0A1L8D8Y4 A0A0L0BV27 A0A1B0FNF6 A0A1B0GGS6 A0A1Q3FWK2 A0A182TIE1 A0A182I6Q4 A0A182V5Q0 A0A182PIJ9 A0A034VNH2 A0A0K8UEF0 W8AMU0 A0A0A1X7P5 A0A0J9RNK4 B4PDV7 B4HW20 B3NEX0 Q9W073 B3M789 A0A2M3ZCC4 A0A2M4C1F1 T1E899 A0A2M4CSC9 A0A182FEM9 A0A2M4ARH0 A0A182IZX6 A0A182L3N7 Q7QIK9 A0A084W132 A0A182WBH6 A0A182N9B6 A0A182RJK2 A0A1W4W7W5 Q2M060 B4GS31 A0A182Y2A9 A0A182T2C6 A0A182JXN1 A0A1Q3F470 Q16NS2 A0A3B0KIH8 B0WLV3 A0A182QAR1 A0A1L8D9C6 A0A1B6L8V2 A0A182U3H7 B4L9P0 A0A1B6I2A6 A0A0M4EBZ8 A0A2J7PX52 A0A182WWH4 A0A1Y1LYA2 A0A067RF78 B4N4H3 R4V237 B4IZY1 B4LDE6 A0A2M3ZD57 A0A2M3ZCX6 D2A630 A0A1B6E2C2 N6U8Y3 J3JU09 U4U676 A0A2M4CH14 A0A1I8NG06 A0A1W4WLZ5 V5GL79 A0A1B6EK55 A0A2H8TNA7 A0A1D2N501 J9K1Z2 A0A2S2NY20 E9FUR6 A0A164YPH7 A0A2R5LGA7 A0A2P6LC32
S4P753 A0A2A4K7L9 A0A1E1W935 A0A2H1VTB1 A0A2A4K777 A0A0K8TQ01 A0A182GQ91 W5JQC7 T1P8Q1 U5EQQ6 A0A1L8D8Y4 A0A0L0BV27 A0A1B0FNF6 A0A1B0GGS6 A0A1Q3FWK2 A0A182TIE1 A0A182I6Q4 A0A182V5Q0 A0A182PIJ9 A0A034VNH2 A0A0K8UEF0 W8AMU0 A0A0A1X7P5 A0A0J9RNK4 B4PDV7 B4HW20 B3NEX0 Q9W073 B3M789 A0A2M3ZCC4 A0A2M4C1F1 T1E899 A0A2M4CSC9 A0A182FEM9 A0A2M4ARH0 A0A182IZX6 A0A182L3N7 Q7QIK9 A0A084W132 A0A182WBH6 A0A182N9B6 A0A182RJK2 A0A1W4W7W5 Q2M060 B4GS31 A0A182Y2A9 A0A182T2C6 A0A182JXN1 A0A1Q3F470 Q16NS2 A0A3B0KIH8 B0WLV3 A0A182QAR1 A0A1L8D9C6 A0A1B6L8V2 A0A182U3H7 B4L9P0 A0A1B6I2A6 A0A0M4EBZ8 A0A2J7PX52 A0A182WWH4 A0A1Y1LYA2 A0A067RF78 B4N4H3 R4V237 B4IZY1 B4LDE6 A0A2M3ZD57 A0A2M3ZCX6 D2A630 A0A1B6E2C2 N6U8Y3 J3JU09 U4U676 A0A2M4CH14 A0A1I8NG06 A0A1W4WLZ5 V5GL79 A0A1B6EK55 A0A2H8TNA7 A0A1D2N501 J9K1Z2 A0A2S2NY20 E9FUR6 A0A164YPH7 A0A2R5LGA7 A0A2P6LC32
Pubmed
19121390
26354079
26227816
22118469
23622113
26369729
+ More
26483478 20920257 23761445 26108605 25348373 24495485 25830018 22936249 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20966253 12364791 14747013 17210077 24438588 15632085 25244985 17510324 28004739 24845553 18362917 19820115 23537049 22516182 25315136 27289101 21292972
26483478 20920257 23761445 26108605 25348373 24495485 25830018 22936249 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20966253 12364791 14747013 17210077 24438588 15632085 25244985 17510324 28004739 24845553 18362917 19820115 23537049 22516182 25315136 27289101 21292972
EMBL
BABH01004176
KQ459582
KPI98936.1
KQ459700
KPJ20353.1
RSAL01000071
+ More
RVE49099.1 JTDY01000595 KOB76536.1 AGBW02007653 OWR54887.1 GAIX01006551 JAA86009.1 NWSH01000086 PCG79763.1 GDQN01007568 JAT83486.1 ODYU01004311 SOQ44049.1 PCG79764.1 GDAI01001131 JAI16472.1 JXUM01015666 KQ560437 KXJ82414.1 ADMH02000413 ETN66617.1 KA645151 AFP59780.1 GANO01004121 JAB55750.1 GFDF01011138 JAV02946.1 JRES01001294 KNC23858.1 CCAG010000676 AJWK01000681 AJWK01000682 AJWK01000683 GFDL01003110 JAV31935.1 APCN01003693 GAKP01014086 JAC44866.1 GDHF01033100 GDHF01027262 JAI19214.1 JAI25052.1 GAMC01020767 GAMC01020766 GAMC01020765 JAB85788.1 GBXI01007381 JAD06911.1 CM002912 KMY96969.1 CM000159 EDW92922.1 CH480817 EDW50135.1 CH954178 EDV50243.1 AE014296 AY119162 AAF47585.1 AAM51022.1 AGB93991.1 CH902618 EDV38750.1 GGFM01005443 MBW26194.1 GGFJ01009988 MBW59129.1 GAMD01002958 JAA98632.1 GGFL01004074 MBW68252.1 GGFK01010052 MBW43373.1 AAAB01008807 EAA04100.4 ATLV01019208 KE525264 KFB43926.1 CH379069 EAL31076.1 CH479188 EDW40566.1 GFDL01012726 JAV22319.1 CH477810 EAT36010.1 OUUW01000009 SPP84891.1 DS233476 DS231990 EDS30154.1 EDS30681.1 AXCN02000559 GFDF01011180 JAV02904.1 GEBQ01019840 JAT20137.1 CH933816 EDW17088.1 GECU01026639 JAS81067.1 CP012525 ALC42871.1 NEVH01020862 PNF20913.1 GEZM01044119 JAV78512.1 KK852507 KDR22427.1 CH964095 EDW79047.1 KC740953 AGM32777.1 CH916366 EDV96753.1 CH940647 EDW68884.1 GGFM01005678 MBW26429.1 GGFM01005656 MBW26407.1 KQ971346 EFA04989.1 GEDC01010343 GEDC01006976 GEDC01005213 JAS26955.1 JAS30322.1 JAS32085.1 APGK01044205 KB741021 ENN75047.1 BT126720 AEE61682.1 KB632003 ERL87823.1 GGFL01000030 MBW64208.1 GALX01007328 JAB61138.1 GECZ01031488 JAS38281.1 GFXV01002953 MBW14758.1 LJIJ01000216 ODN00301.1 ABLF02024009 GGMR01008977 MBY21596.1 GL732525 EFX88775.1 LRGB01000868 KZS15464.1 GGLE01004416 MBY08542.1 MWRG01000423 PRD36147.1
RVE49099.1 JTDY01000595 KOB76536.1 AGBW02007653 OWR54887.1 GAIX01006551 JAA86009.1 NWSH01000086 PCG79763.1 GDQN01007568 JAT83486.1 ODYU01004311 SOQ44049.1 PCG79764.1 GDAI01001131 JAI16472.1 JXUM01015666 KQ560437 KXJ82414.1 ADMH02000413 ETN66617.1 KA645151 AFP59780.1 GANO01004121 JAB55750.1 GFDF01011138 JAV02946.1 JRES01001294 KNC23858.1 CCAG010000676 AJWK01000681 AJWK01000682 AJWK01000683 GFDL01003110 JAV31935.1 APCN01003693 GAKP01014086 JAC44866.1 GDHF01033100 GDHF01027262 JAI19214.1 JAI25052.1 GAMC01020767 GAMC01020766 GAMC01020765 JAB85788.1 GBXI01007381 JAD06911.1 CM002912 KMY96969.1 CM000159 EDW92922.1 CH480817 EDW50135.1 CH954178 EDV50243.1 AE014296 AY119162 AAF47585.1 AAM51022.1 AGB93991.1 CH902618 EDV38750.1 GGFM01005443 MBW26194.1 GGFJ01009988 MBW59129.1 GAMD01002958 JAA98632.1 GGFL01004074 MBW68252.1 GGFK01010052 MBW43373.1 AAAB01008807 EAA04100.4 ATLV01019208 KE525264 KFB43926.1 CH379069 EAL31076.1 CH479188 EDW40566.1 GFDL01012726 JAV22319.1 CH477810 EAT36010.1 OUUW01000009 SPP84891.1 DS233476 DS231990 EDS30154.1 EDS30681.1 AXCN02000559 GFDF01011180 JAV02904.1 GEBQ01019840 JAT20137.1 CH933816 EDW17088.1 GECU01026639 JAS81067.1 CP012525 ALC42871.1 NEVH01020862 PNF20913.1 GEZM01044119 JAV78512.1 KK852507 KDR22427.1 CH964095 EDW79047.1 KC740953 AGM32777.1 CH916366 EDV96753.1 CH940647 EDW68884.1 GGFM01005678 MBW26429.1 GGFM01005656 MBW26407.1 KQ971346 EFA04989.1 GEDC01010343 GEDC01006976 GEDC01005213 JAS26955.1 JAS30322.1 JAS32085.1 APGK01044205 KB741021 ENN75047.1 BT126720 AEE61682.1 KB632003 ERL87823.1 GGFL01000030 MBW64208.1 GALX01007328 JAB61138.1 GECZ01031488 JAS38281.1 GFXV01002953 MBW14758.1 LJIJ01000216 ODN00301.1 ABLF02024009 GGMR01008977 MBY21596.1 GL732525 EFX88775.1 LRGB01000868 KZS15464.1 GGLE01004416 MBY08542.1 MWRG01000423 PRD36147.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000283053
UP000037510
UP000007151
+ More
UP000218220 UP000069940 UP000249989 UP000000673 UP000037069 UP000092444 UP000092461 UP000075902 UP000075840 UP000075903 UP000075885 UP000002282 UP000001292 UP000008711 UP000000803 UP000007801 UP000069272 UP000075880 UP000075882 UP000007062 UP000030765 UP000075920 UP000075884 UP000075900 UP000192221 UP000001819 UP000008744 UP000076408 UP000075901 UP000075881 UP000008820 UP000268350 UP000002320 UP000075886 UP000009192 UP000092553 UP000235965 UP000076407 UP000027135 UP000007798 UP000001070 UP000008792 UP000007266 UP000019118 UP000030742 UP000095301 UP000192223 UP000094527 UP000007819 UP000000305 UP000076858
UP000218220 UP000069940 UP000249989 UP000000673 UP000037069 UP000092444 UP000092461 UP000075902 UP000075840 UP000075903 UP000075885 UP000002282 UP000001292 UP000008711 UP000000803 UP000007801 UP000069272 UP000075880 UP000075882 UP000007062 UP000030765 UP000075920 UP000075884 UP000075900 UP000192221 UP000001819 UP000008744 UP000076408 UP000075901 UP000075881 UP000008820 UP000268350 UP000002320 UP000075886 UP000009192 UP000092553 UP000235965 UP000076407 UP000027135 UP000007798 UP000001070 UP000008792 UP000007266 UP000019118 UP000030742 UP000095301 UP000192223 UP000094527 UP000007819 UP000000305 UP000076858
Pfam
PF05915 DUF872
ProteinModelPortal
H9JU70
A0A194Q049
A0A194RR46
A0A437BFF2
A0A0L7LMT7
A0A212FMD1
+ More
S4P753 A0A2A4K7L9 A0A1E1W935 A0A2H1VTB1 A0A2A4K777 A0A0K8TQ01 A0A182GQ91 W5JQC7 T1P8Q1 U5EQQ6 A0A1L8D8Y4 A0A0L0BV27 A0A1B0FNF6 A0A1B0GGS6 A0A1Q3FWK2 A0A182TIE1 A0A182I6Q4 A0A182V5Q0 A0A182PIJ9 A0A034VNH2 A0A0K8UEF0 W8AMU0 A0A0A1X7P5 A0A0J9RNK4 B4PDV7 B4HW20 B3NEX0 Q9W073 B3M789 A0A2M3ZCC4 A0A2M4C1F1 T1E899 A0A2M4CSC9 A0A182FEM9 A0A2M4ARH0 A0A182IZX6 A0A182L3N7 Q7QIK9 A0A084W132 A0A182WBH6 A0A182N9B6 A0A182RJK2 A0A1W4W7W5 Q2M060 B4GS31 A0A182Y2A9 A0A182T2C6 A0A182JXN1 A0A1Q3F470 Q16NS2 A0A3B0KIH8 B0WLV3 A0A182QAR1 A0A1L8D9C6 A0A1B6L8V2 A0A182U3H7 B4L9P0 A0A1B6I2A6 A0A0M4EBZ8 A0A2J7PX52 A0A182WWH4 A0A1Y1LYA2 A0A067RF78 B4N4H3 R4V237 B4IZY1 B4LDE6 A0A2M3ZD57 A0A2M3ZCX6 D2A630 A0A1B6E2C2 N6U8Y3 J3JU09 U4U676 A0A2M4CH14 A0A1I8NG06 A0A1W4WLZ5 V5GL79 A0A1B6EK55 A0A2H8TNA7 A0A1D2N501 J9K1Z2 A0A2S2NY20 E9FUR6 A0A164YPH7 A0A2R5LGA7 A0A2P6LC32
S4P753 A0A2A4K7L9 A0A1E1W935 A0A2H1VTB1 A0A2A4K777 A0A0K8TQ01 A0A182GQ91 W5JQC7 T1P8Q1 U5EQQ6 A0A1L8D8Y4 A0A0L0BV27 A0A1B0FNF6 A0A1B0GGS6 A0A1Q3FWK2 A0A182TIE1 A0A182I6Q4 A0A182V5Q0 A0A182PIJ9 A0A034VNH2 A0A0K8UEF0 W8AMU0 A0A0A1X7P5 A0A0J9RNK4 B4PDV7 B4HW20 B3NEX0 Q9W073 B3M789 A0A2M3ZCC4 A0A2M4C1F1 T1E899 A0A2M4CSC9 A0A182FEM9 A0A2M4ARH0 A0A182IZX6 A0A182L3N7 Q7QIK9 A0A084W132 A0A182WBH6 A0A182N9B6 A0A182RJK2 A0A1W4W7W5 Q2M060 B4GS31 A0A182Y2A9 A0A182T2C6 A0A182JXN1 A0A1Q3F470 Q16NS2 A0A3B0KIH8 B0WLV3 A0A182QAR1 A0A1L8D9C6 A0A1B6L8V2 A0A182U3H7 B4L9P0 A0A1B6I2A6 A0A0M4EBZ8 A0A2J7PX52 A0A182WWH4 A0A1Y1LYA2 A0A067RF78 B4N4H3 R4V237 B4IZY1 B4LDE6 A0A2M3ZD57 A0A2M3ZCX6 D2A630 A0A1B6E2C2 N6U8Y3 J3JU09 U4U676 A0A2M4CH14 A0A1I8NG06 A0A1W4WLZ5 V5GL79 A0A1B6EK55 A0A2H8TNA7 A0A1D2N501 J9K1Z2 A0A2S2NY20 E9FUR6 A0A164YPH7 A0A2R5LGA7 A0A2P6LC32
Ontologies
GO
PANTHER
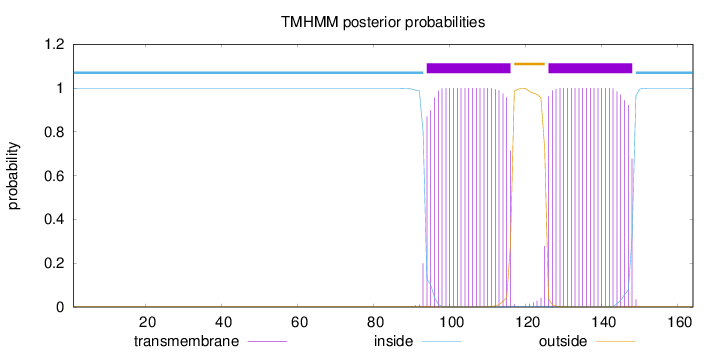
Topology
Length:
164
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
45.44336
Exp number, first 60 AAs:
0
Total prob of N-in:
0.99831
inside
1 - 93
TMhelix
94 - 116
outside
117 - 125
TMhelix
126 - 148
inside
149 - 164
Population Genetic Test Statistics
Pi
292.977454
Theta
236.433311
Tajima's D
0.659501
CLR
0.149735
CSRT
0.562121893905305
Interpretation
Uncertain
