Gene
KWMTBOMO09422
Pre Gene Modal
BGIBMGA013083
Annotation
PREDICTED:_RNA_polymerase_II-associated_protein_1_isoform_X2_[Bombyx_mori]
Full name
Anoctamin
Location in the cell
Nuclear Reliability : 3.995
Sequence
CDS
ATGTTGAGACGACCAACACCAGGTGAAAATGAAGAAGATATTTTAAAAATGCAAGAAGAGTTCCTTCGCAAGAAGAGGCAGAATCAGAACCTGCAGCCGGCAGCACAAGTCGTTAACTTAAGAACTGAACCAATCAAAAGACCAAATACGAATGAACAAGAAATCCGTAAGCCGTCAAAGTACGCACAAAGTAAAGGTCTTAAGAATCATTCCGAGAAACGTTCCAAAAGTAATGACACAGTTGGAAACTCCGTCATAACTGATGTTTATGAAAAAAATACAGAAAAACCAGTCGAAAAGAAACTTGACGGCGAAGATGACAAGGTTTACTATCCAACAATACTTCCATTTGTATTAGGGGACATTGTCGAAAAGAATTTAGATACTGTAGTTGAGTTTGATTATAAGCCAATGCCACCAGAAGGATTTCCAGTTGTAAGAAAACAAAATAAAGACTCTAAGGCCACTAAGTATATATATCAGAAAAATTTAAATAAAATTAAAAAAAATATAGTTGATAAAATGGATGTAGATGAACCATCATGCTCAAATAATAAACAAAACACCATAAACCTGCCAGATAGAAGTTACATATTGACTTCAGAAAACTCTGATGACATCCACAATGAAAATATAAAAATTCTAAGTAAGATGTCCGAAAAAGAAATTTTAGAAGAACAGCAAAAACTATTCTCTAATTTAGATCCTAAAGTGGTTGACTTTATTAAATCACGGAGAATGCAAAATGAGAACTCAACTCGACAAACACTAAAGTTGAATGAAAGAGATGATATAAATAAGCTAGCTACTGTGAACCAAAATGTAGAAATGGACACAAGCACTCAGGAAATTGAAAATGATACATTATGGGAGAATGATGTTCTTTCTCACCCTGAAATACAAAATTGGCTGCATTTCGACTCGCTAGAGAAAGACAAGCTTCAATGGATGGAAGGACTTGATGAAAGTAAAAACATTAAGCCTGATGAGCCTTATGAAGCGAGGTTTGATTTCAAAGGATATTTACTACCTTATACAATGGAATACACTGATATGACTAAAACTTTATATCACCACGGTGAAGAGCCGCACCGTCCTGGGTATACTATCACAGAGTTAATTGAGTTGACACGGTCAACTATTACACAGCAGAGAGTTATGGCCTTAAATACAATTGCTGGTATCCTCCAGTATCATAGTGCTGGAACTTACAAAGACATTATAGAAATACCATTGAGCAAGTTGTTTTTCATCATAAGAATTGCAATGGATGAAAATAAAGTTATAATTCTTGAACCTGCTTTGAAAGCAATGAGAAATTTAGTGTACAACAAGTTAGATGAAGCGTCGCTCGATGCATTGATAGGATTTGAAGAAGGAAACCTGCAGCCTTGCTTTGAAAATGATAAATCAGAAATAGAGGAACTAGAATCAAAAGAAGCAGAGTTAAAAGACTTTCATTTAGCAGAGATTGATTTAATTGCAGCCCTATTAAGAACAGATATACTGCAAAGGATATACTATATATTAGAGTCAATAAAACCAAGCTTCAACTGTGTACAGTATTGCTTACAGATATTAACAAGACTAGGTAGAGATTCTGTCGAGACTGCCAAAAGAATCTGTGAAATGGAGCACCTCATGAACTCTATAATAACAAATTTTGTTCCGGTGACAGGTGTCAATTTTGTCTTCAAACCACAAGTTGTATATAATGGTAAACCAGTTTTAGCAGCTATAAAGCTACTAAGAGTTCTAAGTCTACAAACTGAAGAAATTTGCAATATTTTACTAATGAAATACAATATTTTAGCACCAATTTCTGTGTATCTGAGTTCAGCAGTTGACAGAAGTTATGGCTTAAGAATACAGACTGAGTCATATTGTATACTATCAAATTTACTTTATTTTGGACTAAGCTATGATAGTGCCATTTCATTGTGTCCAGTCATACTGACAGCTTTATACAAACACGTTCAGGGTACAGACATTAATGTGGAAGCTTCTGTGTTGGCAGCAACACATGCAGCTGTGGTCTTGCAGCTGATAAATAACATACTGAAACATAATTTAAATGATTTTCTTACTTTCAAGACTGAAGTTTATAAGTTGTTGAAGGAAGGGCTATTGAAATGGTTTGGTCAGTTATCGCAAATGGAACATTATACATGTGCGCATCTGAGAATTTTATGCTCGGCTTTAGATTGCTGTTATACTGTTATTGAAATTGAACAATTGCCTGTGAAGTTCGTGAGTGATGCCCTGAAACAACTGTCTTATTCTAAAGGATTTCAAAATGTGATTAACAACCTTAGTTCCTGCTCTAATTTGCTCTCAAAAATTGAAAATAAAGATTTGTATTCAGTTAAGAACTTAATAAGTCTAAAATGCACTGTCATTGATTCTGCTCAAAAAGTTTTACCAATACTGAAAGTTGCATCTCCCATACCTCTGCTGATGTCACTTTTCAAATTACTGACAATAGTCAATCAAAAAGAAATCTCTAGAATATTCTTAGACCATGTTTGGAACTATTTGCAAAGATTATCTAAATGTGTTCCTAGTCTTTCAGATAATTGGTTCACTAGAATGGAAACTAATTTTGTTTTTTACGTTATGAAATTATCTGTTATGTGTAATGTTGATGAAAGCAAAAAGGATGTAATGTATGCAGTAGCTAACAAATTGTGTTACACATTGAGGGTAGATAAAAAATATGAACTAGATTTCTTATTTGAAAGTATCGTTTTTAACAAAGATTGGTTCACGGCCGAAAGGTTGTTAAATCTTGTCAATTTGACTGATGCAGACGGATTCTCAAGAGCTTTAACTGCCATAGAAGACATACGACAATGTTACCGTAGAGTTATTAACTTGAATTACAAAGATACAGGTGCTAATGTTGTGTTAAGAAAATGGATGGAGCCAATTTTGCCAAGAGACTGGATTTATTTGCCAATTTTGATGTTATACAGTAAAAATCAGGAGGCGGAGTCGATTCCCCATGTTTCCGGAGATCATGCCCAGAAAATGTCAGCGCAAATAGCGACCGAAAAGGAATTTATAATAACATGTAGCTTAGAATGGATATTGTTTAACGAACTCTGTTTTTCTGATTTAGTCAATGATATTGATGTAACAGATAGATTTTGTCGAATCATGTGCGTGTTTTTATGTGACAACTCGTTATTCTTGCAACCAAAAATTAGAATGTTACTGCAGAAATGCACCAATGTCCTTTTCAAAAAGAAATCAGCCTTCAATTTCGATAAACAATTAACGGGCTTGAACAACTTCCAAGATTTCTACACACAGTTTCTAGAACAATTCCAAGCGGTCAGCTATGGAGATGACGTGTTTTCTGCATGTGTATTGGTGCCTTTAGCTCAAAAGCATAATGTTAAATGGAGGAAATTGATATGGTCGGAATATGCTGGATGTCTCCGGGCGTTAGATTGCCCGGAACAACTACTTTGTTATGAAGTAAATGAGTATCTTTATCCAGAAGAGACGGACGAATCTTTGATTAAATCTTATCATCGCGCGCTATCCTGTAATTTACTACGACCGGATACAATAATGCATAAAATTGCGAGTCATCACGTGCACATATTCAAGAATCGATCTCAGATACAAAACACTTGA
Protein
MLRRPTPGENEEDILKMQEEFLRKKRQNQNLQPAAQVVNLRTEPIKRPNTNEQEIRKPSKYAQSKGLKNHSEKRSKSNDTVGNSVITDVYEKNTEKPVEKKLDGEDDKVYYPTILPFVLGDIVEKNLDTVVEFDYKPMPPEGFPVVRKQNKDSKATKYIYQKNLNKIKKNIVDKMDVDEPSCSNNKQNTINLPDRSYILTSENSDDIHNENIKILSKMSEKEILEEQQKLFSNLDPKVVDFIKSRRMQNENSTRQTLKLNERDDINKLATVNQNVEMDTSTQEIENDTLWENDVLSHPEIQNWLHFDSLEKDKLQWMEGLDESKNIKPDEPYEARFDFKGYLLPYTMEYTDMTKTLYHHGEEPHRPGYTITELIELTRSTITQQRVMALNTIAGILQYHSAGTYKDIIEIPLSKLFFIIRIAMDENKVIILEPALKAMRNLVYNKLDEASLDALIGFEEGNLQPCFENDKSEIEELESKEAELKDFHLAEIDLIAALLRTDILQRIYYILESIKPSFNCVQYCLQILTRLGRDSVETAKRICEMEHLMNSIITNFVPVTGVNFVFKPQVVYNGKPVLAAIKLLRVLSLQTEEICNILLMKYNILAPISVYLSSAVDRSYGLRIQTESYCILSNLLYFGLSYDSAISLCPVILTALYKHVQGTDINVEASVLAATHAAVVLQLINNILKHNLNDFLTFKTEVYKLLKEGLLKWFGQLSQMEHYTCAHLRILCSALDCCYTVIEIEQLPVKFVSDALKQLSYSKGFQNVINNLSSCSNLLSKIENKDLYSVKNLISLKCTVIDSAQKVLPILKVASPIPLLMSLFKLLTIVNQKEISRIFLDHVWNYLQRLSKCVPSLSDNWFTRMETNFVFYVMKLSVMCNVDESKKDVMYAVANKLCYTLRVDKKYELDFLFESIVFNKDWFTAERLLNLVNLTDADGFSRALTAIEDIRQCYRRVINLNYKDTGANVVLRKWMEPILPRDWIYLPILMLYSKNQEAESIPHVSGDHAQKMSAQIATEKEFIITCSLEWILFNELCFSDLVNDIDVTDRFCRIMCVFLCDNSLFLQPKIRMLLQKCTNVLFKKKSAFNFDKQLTGLNNFQDFYTQFLEQFQAVSYGDDVFSACVLVPLAQKHNVKWRKLIWSEYAGCLRALDCPEQLLCYEVNEYLYPEETDESLIKSYHRALSCNLLRPDTIMHKIASHHVHIFKNRSQIQNT
Summary
Similarity
Belongs to the anoctamin family.
Uniprot
H9JU71
A0A2H1VT93
A0A2A4K7X4
A0A0L7LMF0
A0A194RSN1
A0A194Q0V0
+ More
A0A1W4WTY0 D6WZF8 A0A1Y1JZ13 A0A1Y1JVY9 A0A1Y1K3Q0 U4UNT9 N6U467 A0A2J7R5R9 A0A1I8MD34 A0A067R0A2 A0A1I8Q6B7 A0A0L0BYS7 A0A088AD45 A0A0K8V393 A0A2A3EM84 W8BVK7 A0A182PGT8 A0A0N0U3Y5 A0A195AUE5 A0A1A9ZVS9 K7IMJ9 A0A154PES7 A0A1A9URM2 A0A158NNA2 A0A1B6CGL5 A0A3L8DHG6 F4WLI3 A0A026WVF4 A0A1B0CE78 A0A182IZE9 A0A1B0BLP3 E2B3G7 A0A1B0G4L8 A0A232FE49 A0A195FWP2 A0A2P8YY09 A0A0L7QWW2 A0A1J1IC21 A0A1A9XJE3 A0A1B6LT30 A0A1S4G730 A0A0T6ATZ4 A0A224XIZ1 B4QR95 A0A151XA83 A0A2M4CJG7 A0A1B6L9P3 A0A069DXF9 E2AJ22 E9IUF0 B4N3I5 A0A0V0G857 A0A0J7KRX8 A0A195CRA7 A0A0A9W2X6 A0A146L171 K1R846 A0A3P9M437 A0A3P9KNS2 A0A3P9J5F7 A0A3P9J5T1 A0A3B3I2J7 H2MKR8 A0A3B3Y1K6 A0A3B3VQR2 E0VJE1 A0A315V308 A0A2D0RPZ9 A0A087XIT2 A0A3B3BN04 A0A336LKS0 A0A1I8Q692 A0A0C9RQ85 A0A3Q1AX48 A0A3B4CXT0 H2UAQ7 A0A3B4WRY9 U3JU85 A0A3Q1JYT9 A0A091QNS0 A0A3Q1JYV1 A0A437C3X8 H3DIW6 F1NLT1 A0A093EES5 A0A091SP90 A0A3B4V5G3
A0A1W4WTY0 D6WZF8 A0A1Y1JZ13 A0A1Y1JVY9 A0A1Y1K3Q0 U4UNT9 N6U467 A0A2J7R5R9 A0A1I8MD34 A0A067R0A2 A0A1I8Q6B7 A0A0L0BYS7 A0A088AD45 A0A0K8V393 A0A2A3EM84 W8BVK7 A0A182PGT8 A0A0N0U3Y5 A0A195AUE5 A0A1A9ZVS9 K7IMJ9 A0A154PES7 A0A1A9URM2 A0A158NNA2 A0A1B6CGL5 A0A3L8DHG6 F4WLI3 A0A026WVF4 A0A1B0CE78 A0A182IZE9 A0A1B0BLP3 E2B3G7 A0A1B0G4L8 A0A232FE49 A0A195FWP2 A0A2P8YY09 A0A0L7QWW2 A0A1J1IC21 A0A1A9XJE3 A0A1B6LT30 A0A1S4G730 A0A0T6ATZ4 A0A224XIZ1 B4QR95 A0A151XA83 A0A2M4CJG7 A0A1B6L9P3 A0A069DXF9 E2AJ22 E9IUF0 B4N3I5 A0A0V0G857 A0A0J7KRX8 A0A195CRA7 A0A0A9W2X6 A0A146L171 K1R846 A0A3P9M437 A0A3P9KNS2 A0A3P9J5F7 A0A3P9J5T1 A0A3B3I2J7 H2MKR8 A0A3B3Y1K6 A0A3B3VQR2 E0VJE1 A0A315V308 A0A2D0RPZ9 A0A087XIT2 A0A3B3BN04 A0A336LKS0 A0A1I8Q692 A0A0C9RQ85 A0A3Q1AX48 A0A3B4CXT0 H2UAQ7 A0A3B4WRY9 U3JU85 A0A3Q1JYT9 A0A091QNS0 A0A3Q1JYV1 A0A437C3X8 H3DIW6 F1NLT1 A0A093EES5 A0A091SP90 A0A3B4V5G3
Pubmed
19121390
26227816
26354079
18362917
19820115
28004739
+ More
23537049 25315136 24845553 26108605 24495485 20075255 21347285 30249741 21719571 24508170 20798317 28648823 29403074 17510324 17994087 22936249 26334808 21282665 25401762 26823975 22992520 17554307 20566863 29703783 29451363 21551351 15496914 15592404
23537049 25315136 24845553 26108605 24495485 20075255 21347285 30249741 21719571 24508170 20798317 28648823 29403074 17510324 17994087 22936249 26334808 21282665 25401762 26823975 22992520 17554307 20566863 29703783 29451363 21551351 15496914 15592404
EMBL
BABH01004178
ODYU01004311
SOQ44047.1
NWSH01000086
PCG79760.1
JTDY01000595
+ More
KOB76534.1 KQ459700 KPJ20355.1 KQ459582 KPI98938.1 KQ971372 EFA10420.1 GEZM01099127 JAV53478.1 GEZM01099126 JAV53479.1 GEZM01099128 JAV53477.1 KB632381 ERL94148.1 APGK01043331 APGK01043332 APGK01043333 KB741014 ENN75421.1 NEVH01006990 PNF36178.1 KK852832 KDR15317.1 JRES01001238 KNC24389.1 GDHF01019249 JAI33065.1 KZ288220 PBC32151.1 GAMC01013361 JAB93194.1 KQ435878 KOX70008.1 KQ976738 KYM75858.1 KQ434889 KZC10341.1 ADTU01021196 GEDC01024759 JAS12539.1 QOIP01000008 RLU19890.1 GL888208 EGI64919.1 KK107109 EZA59089.1 AJWK01008657 JXJN01016527 GL445346 EFN89719.1 CCAG010006129 NNAY01000356 OXU28942.1 KQ981208 KYN44737.1 PYGN01000293 PSN49130.1 KQ414706 KOC63085.1 CVRI01000047 CRK97831.1 GEBQ01013170 JAT26807.1 LJIG01022847 KRT78413.1 GFTR01008425 JAW08001.1 CM000363 CM002912 EDX10225.1 KMY99221.1 KQ982351 KYQ57281.1 GGFL01001133 MBW65311.1 GEBQ01019557 JAT20420.1 GBGD01000304 JAC88585.1 GL439932 EFN66554.1 GL765974 EFZ15790.1 CH964095 EDW79190.1 GECL01001770 JAP04354.1 LBMM01003800 KMQ93122.1 KQ977372 KYN03273.1 GBHO01041460 JAG02144.1 GDHC01016316 JAQ02313.1 JH818024 EKC39839.1 DS235222 EEB13497.1 NHOQ01002481 PWA16478.1 AYCK01008751 UFQT01000044 SSX18794.1 GBYB01010675 JAG80442.1 AGTO01021192 KK800801 KFQ28569.1 CM012458 RVE57278.1 AADN05000303 KL461090 KFV13001.1 KK930350 KFQ44847.1
KOB76534.1 KQ459700 KPJ20355.1 KQ459582 KPI98938.1 KQ971372 EFA10420.1 GEZM01099127 JAV53478.1 GEZM01099126 JAV53479.1 GEZM01099128 JAV53477.1 KB632381 ERL94148.1 APGK01043331 APGK01043332 APGK01043333 KB741014 ENN75421.1 NEVH01006990 PNF36178.1 KK852832 KDR15317.1 JRES01001238 KNC24389.1 GDHF01019249 JAI33065.1 KZ288220 PBC32151.1 GAMC01013361 JAB93194.1 KQ435878 KOX70008.1 KQ976738 KYM75858.1 KQ434889 KZC10341.1 ADTU01021196 GEDC01024759 JAS12539.1 QOIP01000008 RLU19890.1 GL888208 EGI64919.1 KK107109 EZA59089.1 AJWK01008657 JXJN01016527 GL445346 EFN89719.1 CCAG010006129 NNAY01000356 OXU28942.1 KQ981208 KYN44737.1 PYGN01000293 PSN49130.1 KQ414706 KOC63085.1 CVRI01000047 CRK97831.1 GEBQ01013170 JAT26807.1 LJIG01022847 KRT78413.1 GFTR01008425 JAW08001.1 CM000363 CM002912 EDX10225.1 KMY99221.1 KQ982351 KYQ57281.1 GGFL01001133 MBW65311.1 GEBQ01019557 JAT20420.1 GBGD01000304 JAC88585.1 GL439932 EFN66554.1 GL765974 EFZ15790.1 CH964095 EDW79190.1 GECL01001770 JAP04354.1 LBMM01003800 KMQ93122.1 KQ977372 KYN03273.1 GBHO01041460 JAG02144.1 GDHC01016316 JAQ02313.1 JH818024 EKC39839.1 DS235222 EEB13497.1 NHOQ01002481 PWA16478.1 AYCK01008751 UFQT01000044 SSX18794.1 GBYB01010675 JAG80442.1 AGTO01021192 KK800801 KFQ28569.1 CM012458 RVE57278.1 AADN05000303 KL461090 KFV13001.1 KK930350 KFQ44847.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053240
UP000053268
UP000192223
+ More
UP000007266 UP000030742 UP000019118 UP000235965 UP000095301 UP000027135 UP000095300 UP000037069 UP000005203 UP000242457 UP000075885 UP000053105 UP000078540 UP000092445 UP000002358 UP000076502 UP000078200 UP000005205 UP000279307 UP000007755 UP000053097 UP000092461 UP000075880 UP000092460 UP000008237 UP000092444 UP000215335 UP000078541 UP000245037 UP000053825 UP000183832 UP000092443 UP000000304 UP000075809 UP000000311 UP000007798 UP000036403 UP000078542 UP000005408 UP000265180 UP000265200 UP000001038 UP000261480 UP000261500 UP000009046 UP000221080 UP000028760 UP000261560 UP000257160 UP000261440 UP000005226 UP000261360 UP000016665 UP000265040 UP000007303 UP000000539 UP000261420
UP000007266 UP000030742 UP000019118 UP000235965 UP000095301 UP000027135 UP000095300 UP000037069 UP000005203 UP000242457 UP000075885 UP000053105 UP000078540 UP000092445 UP000002358 UP000076502 UP000078200 UP000005205 UP000279307 UP000007755 UP000053097 UP000092461 UP000075880 UP000092460 UP000008237 UP000092444 UP000215335 UP000078541 UP000245037 UP000053825 UP000183832 UP000092443 UP000000304 UP000075809 UP000000311 UP000007798 UP000036403 UP000078542 UP000005408 UP000265180 UP000265200 UP000001038 UP000261480 UP000261500 UP000009046 UP000221080 UP000028760 UP000261560 UP000257160 UP000261440 UP000005226 UP000261360 UP000016665 UP000265040 UP000007303 UP000000539 UP000261420
PRIDE
Pfam
Interpro
IPR013930
RNA_pol_II_AP1_N
+ More
IPR039913 RPAP1/Rba50
IPR013929 RNA_pol_II_AP1_C
IPR016024 ARM-type_fold
IPR011989 ARM-like
IPR004895 Prenylated_rab_accept_PRA1
IPR032394 Anoct_dimer
IPR007632 Anoctamin
IPR001254 Trypsin_dom
IPR001314 Peptidase_S1A
IPR009003 Peptidase_S1_PA
IPR013604 7TM_chemorcpt
IPR039913 RPAP1/Rba50
IPR013929 RNA_pol_II_AP1_C
IPR016024 ARM-type_fold
IPR011989 ARM-like
IPR004895 Prenylated_rab_accept_PRA1
IPR032394 Anoct_dimer
IPR007632 Anoctamin
IPR001254 Trypsin_dom
IPR001314 Peptidase_S1A
IPR009003 Peptidase_S1_PA
IPR013604 7TM_chemorcpt
Gene 3D
CDD
ProteinModelPortal
H9JU71
A0A2H1VT93
A0A2A4K7X4
A0A0L7LMF0
A0A194RSN1
A0A194Q0V0
+ More
A0A1W4WTY0 D6WZF8 A0A1Y1JZ13 A0A1Y1JVY9 A0A1Y1K3Q0 U4UNT9 N6U467 A0A2J7R5R9 A0A1I8MD34 A0A067R0A2 A0A1I8Q6B7 A0A0L0BYS7 A0A088AD45 A0A0K8V393 A0A2A3EM84 W8BVK7 A0A182PGT8 A0A0N0U3Y5 A0A195AUE5 A0A1A9ZVS9 K7IMJ9 A0A154PES7 A0A1A9URM2 A0A158NNA2 A0A1B6CGL5 A0A3L8DHG6 F4WLI3 A0A026WVF4 A0A1B0CE78 A0A182IZE9 A0A1B0BLP3 E2B3G7 A0A1B0G4L8 A0A232FE49 A0A195FWP2 A0A2P8YY09 A0A0L7QWW2 A0A1J1IC21 A0A1A9XJE3 A0A1B6LT30 A0A1S4G730 A0A0T6ATZ4 A0A224XIZ1 B4QR95 A0A151XA83 A0A2M4CJG7 A0A1B6L9P3 A0A069DXF9 E2AJ22 E9IUF0 B4N3I5 A0A0V0G857 A0A0J7KRX8 A0A195CRA7 A0A0A9W2X6 A0A146L171 K1R846 A0A3P9M437 A0A3P9KNS2 A0A3P9J5F7 A0A3P9J5T1 A0A3B3I2J7 H2MKR8 A0A3B3Y1K6 A0A3B3VQR2 E0VJE1 A0A315V308 A0A2D0RPZ9 A0A087XIT2 A0A3B3BN04 A0A336LKS0 A0A1I8Q692 A0A0C9RQ85 A0A3Q1AX48 A0A3B4CXT0 H2UAQ7 A0A3B4WRY9 U3JU85 A0A3Q1JYT9 A0A091QNS0 A0A3Q1JYV1 A0A437C3X8 H3DIW6 F1NLT1 A0A093EES5 A0A091SP90 A0A3B4V5G3
A0A1W4WTY0 D6WZF8 A0A1Y1JZ13 A0A1Y1JVY9 A0A1Y1K3Q0 U4UNT9 N6U467 A0A2J7R5R9 A0A1I8MD34 A0A067R0A2 A0A1I8Q6B7 A0A0L0BYS7 A0A088AD45 A0A0K8V393 A0A2A3EM84 W8BVK7 A0A182PGT8 A0A0N0U3Y5 A0A195AUE5 A0A1A9ZVS9 K7IMJ9 A0A154PES7 A0A1A9URM2 A0A158NNA2 A0A1B6CGL5 A0A3L8DHG6 F4WLI3 A0A026WVF4 A0A1B0CE78 A0A182IZE9 A0A1B0BLP3 E2B3G7 A0A1B0G4L8 A0A232FE49 A0A195FWP2 A0A2P8YY09 A0A0L7QWW2 A0A1J1IC21 A0A1A9XJE3 A0A1B6LT30 A0A1S4G730 A0A0T6ATZ4 A0A224XIZ1 B4QR95 A0A151XA83 A0A2M4CJG7 A0A1B6L9P3 A0A069DXF9 E2AJ22 E9IUF0 B4N3I5 A0A0V0G857 A0A0J7KRX8 A0A195CRA7 A0A0A9W2X6 A0A146L171 K1R846 A0A3P9M437 A0A3P9KNS2 A0A3P9J5F7 A0A3P9J5T1 A0A3B3I2J7 H2MKR8 A0A3B3Y1K6 A0A3B3VQR2 E0VJE1 A0A315V308 A0A2D0RPZ9 A0A087XIT2 A0A3B3BN04 A0A336LKS0 A0A1I8Q692 A0A0C9RQ85 A0A3Q1AX48 A0A3B4CXT0 H2UAQ7 A0A3B4WRY9 U3JU85 A0A3Q1JYT9 A0A091QNS0 A0A3Q1JYV1 A0A437C3X8 H3DIW6 F1NLT1 A0A093EES5 A0A091SP90 A0A3B4V5G3
Ontologies
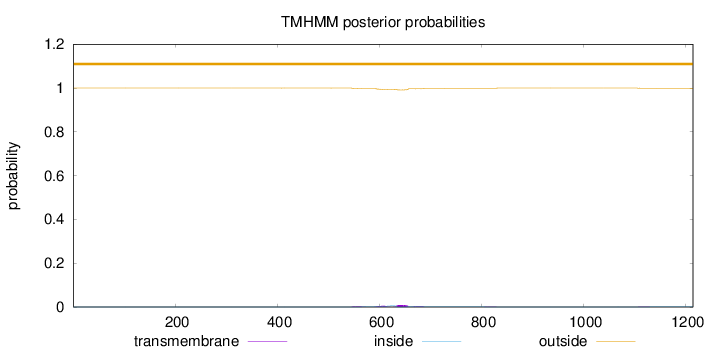
Topology
Subcellular location
Membrane
Length:
1214
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.424370000000002
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00020
outside
1 - 1214
Population Genetic Test Statistics
Pi
163.599172
Theta
175.584325
Tajima's D
-0.232861
CLR
136.021351
CSRT
0.310084495775211
Interpretation
Uncertain
