Gene
KWMTBOMO09418
Pre Gene Modal
BGIBMGA013084
Annotation
PREDICTED:_cell_division_control_protein_45_homolog_[Papilio_machaon]
Full name
Cell division control protein 45 homolog
Alternative Name
PORC-PI-1
Location in the cell
Cytoplasmic Reliability : 1.292 PlasmaMembrane Reliability : 1.157
Sequence
CDS
ATGTTTATAGAGGACCTCAGAAATGACTTTTACAATCTCCTTTTAGGCAATCGCGTGTTATTATTAGTGCATTATGATGTAGATGCAATATGTACATGTAAAATACTTCAAGGATTATTCAAGAGTGATAATATATCATATACGCTAGTGCCGGTGGGGGGTATTTCTGAATTGAAGACTGCTTACGAAGAAAACAATGAAGAAGTCAAGTACGTAGTGTTGGTGAACTGCGGAGGTACTATAGACCTGGTCGATATCTTACAGCCCGAGGAGGATGTGGTATTTTTTGTGCTGGACTCCCATAAACCGACTGATGTCTGCAATGTTTATAGCGATGGACAGGTGAGACTGGTGTACCGTGATGAAGAAGAGAACATTCCTAATTTTGATGATATATTCAGAGATGATGATGATGAGAACGAAGAAGAAATGCCAAGTGGTCGAGAAGACCTAGAAGCTTTTGTCGAGAGACGGCGTCAACGTAGAGAATGGGAAACAAAACGAAACGATCTTATGTTTAACTACACTCAATTCTCATATTATGGTAAACCGAGTGCTTGCATAGCAGTAGAGCTGGCCTGGCTGCTATGTCGCAGCGGAGTGGAGTACACGTGGGCCGGATGCATAGCTGTCAGTGCGTATGCTGCCACTCACACACGCTCCCCTGCACAAGCTCTGCTTGATGCACATGCGCTGCATCGACATCTCGCCGCGCAACTACCTTCACATGGTGCTTCAGTAAGCATGGAAAAGGATGCGTTCCTGCCACTCTACCGGACCTGGTCTTTGGAGAATGCGTTGAAGTATTCTCCAGTTGTGGCTCCTCATCTTCGTCTTCATTCACTGGCTGGTGCTGACAGATTGAGGCAGCTATTGGCAGATATGGGGATCCCGCTGCAGCAGGCCCGCCAGGCCTACCGCTCCATGGACGTGGAGCTGCGGCGCTCGCTGCTGGACGCGCTGGACGCCGCCGCCGACAAGCACCACCTGCCGCGCCTCACGCACGCCACCTACCTCCTGCGCCGCGACCACTGCGCGCCGCTCGCCGCACTCGACTGCGTCTACTGCGTCATGGCGCTGCTCGAGCTCGAAGGCCCCGGCGGGTTCCAGCTGGCGCTGGCGTCGGTGGACACGTCGAGCGCGGAGTGCGAGGCCCGGCAGCGCGGGCTGGGCGCGGCGCGGCGCGGGCTGGCGGCGGCGGCGGCGCTGGCGGGCGCCACGCTGGCGGCGCGCGCGCTGCACCACGCCGGGCCCTTCGCGCACTTCACCATCAGCGAGGGCGCGGCGGAGTGCGAGTGGGGTCCGTGGTGGCTGGCGGTGTGCGCGCGGTGGGTGGGCGCGGGCGCGGGGCGGGGGGCGGCCGCCGTGCTGGTGGCGGCGCCCCGGGGCCGCACCGTGCTGCTGTGCGGGGTGCCGCCGCCGCAGCGGACTGACACCAGGAATCTGTTCGGGGCGGCGTTCGAGCAGGCGGCGGCCAAGTGCGGCGCCTCCGTGTCGCTCGACCACTTCGACTCCGCCGTGGTGACGCTGCCCGCCGCGCAGCGCCCGCAGTTCCTGGACGCGCTCACCGCGCTGCTGGCCTGA
Protein
MFIEDLRNDFYNLLLGNRVLLLVHYDVDAICTCKILQGLFKSDNISYTLVPVGGISELKTAYEENNEEVKYVVLVNCGGTIDLVDILQPEEDVVFFVLDSHKPTDVCNVYSDGQVRLVYRDEEENIPNFDDIFRDDDDENEEEMPSGREDLEAFVERRRQRREWETKRNDLMFNYTQFSYYGKPSACIAVELAWLLCRSGVEYTWAGCIAVSAYAATHTRSPAQALLDAHALHRHLAAQLPSHGASVSMEKDAFLPLYRTWSLENALKYSPVVAPHLRLHSLAGADRLRQLLADMGIPLQQARQAYRSMDVELRRSLLDALDAAADKHHLPRLTHATYLLRRDHCAPLAALDCVYCVMALLELEGPGGFQLALASVDTSSAECEARQRGLGAARRGLAAAAALAGATLAARALHHAGPFAHFTISEGAAECEWGPWWLAVCARWVGAGAGRGAAAVLVAAPRGRTVLLCGVPPPQRTDTRNLFGAAFEQAAAKCGASVSLDHFDSAVVTLPAAQRPQFLDALTALLA
Summary
Subunit
Associated with ORC2. Interacts with HELB.
Associated with ORC2. Interacts with HELB (PubMed:25933514).
Associated with ORC2. Interacts with HELB (PubMed:25933514).
Similarity
Belongs to the CDC45 family.
Keywords
Cell cycle
Complete proteome
DNA replication
Nucleus
Phosphoprotein
Reference proteome
3D-structure
Alternative splicing
Cytoplasm
Disease mutation
Dwarfism
Polymorphism
Feature
chain Cell division control protein 45 homolog
splice variant In isoform 2.
sequence variant In MGORS7; decreased protein level; dbSNP:rs879255633.
splice variant In isoform 2.
sequence variant In MGORS7; decreased protein level; dbSNP:rs879255633.
Uniprot
H9JU72
A0A2H1VKQ4
A0A2A4K7M8
A0A194Q1R9
A0A194RVZ2
T1HHG2
+ More
A0A023F0J3 A0A067R9J8 A0A2R7WKB3 A0A224XN74 A0A2J7RBG6 A0A1Y1LJK7 A0A2S2R8N5 A0A1W4X9Z5 A0A1B6K275 A0A2S2NN53 A0A182GR65 A0A2H8TQS0 A0A232FJQ4 A0A1Y1LES8 J9K5U7 Q16EN6 A0A026VYI2 A0A182Y9K9 A0A3L8DQJ9 A0A0M9AAS8 E2BK22 Q170F3 A0A0C9QZG8 A0A182M445 A0A1B0CPX3 J9K7X0 A0A195BKW3 A0A154PU39 A0A2S2PQW2 A0A158P3W2 A0A1B0D3Z1 F4WKV8 A0A310SCM5 A0A087ZR06 D2A4C4 A0A151WV55 A0A336L5R6 A0A1A9WVN8 A0A1B0BDI7 A0A1A9ZKR9 A0A2H8TX98 A0A0L7QLB8 A0A182Q3S6 A0A2A3E1X5 A0A1A9YME8 A0A3R7M7E1 E1BYS7 A0A2U4ALI7 R7VUU8 A0A3Q2UBE9 H0ZDT9 A0A182FV47 A0A087V099 A0A341CYT0 A4FV47 A0A2Y9FDE4 A0A384BAJ1 A0A3Q7VW59 H9GZX0 Q3UI99 Q9Z1X9 A0A0A9YQW4 K7GSB7 F1MQQ0 Q5E9W1 A0A1U7TNB8 A0A3Q7U1N8 A0A061I7M6 Q4U3P7 G3QNC9 A0A2R9A6H4 A0A2Y9LBD2 A0A2Y9H4H2 A0A2U3YXH3 O75419 A0A3P4LWF5 A0A2I2Y746 A0A2R9A140 L8I6K2 A0A3Q7MZ45 K7A671 A0A2K5IJU4 B4GTR9 A0A2K6SB67 F7IIN6 A0A2Y9J0S9 A0A2K6P4W2
A0A023F0J3 A0A067R9J8 A0A2R7WKB3 A0A224XN74 A0A2J7RBG6 A0A1Y1LJK7 A0A2S2R8N5 A0A1W4X9Z5 A0A1B6K275 A0A2S2NN53 A0A182GR65 A0A2H8TQS0 A0A232FJQ4 A0A1Y1LES8 J9K5U7 Q16EN6 A0A026VYI2 A0A182Y9K9 A0A3L8DQJ9 A0A0M9AAS8 E2BK22 Q170F3 A0A0C9QZG8 A0A182M445 A0A1B0CPX3 J9K7X0 A0A195BKW3 A0A154PU39 A0A2S2PQW2 A0A158P3W2 A0A1B0D3Z1 F4WKV8 A0A310SCM5 A0A087ZR06 D2A4C4 A0A151WV55 A0A336L5R6 A0A1A9WVN8 A0A1B0BDI7 A0A1A9ZKR9 A0A2H8TX98 A0A0L7QLB8 A0A182Q3S6 A0A2A3E1X5 A0A1A9YME8 A0A3R7M7E1 E1BYS7 A0A2U4ALI7 R7VUU8 A0A3Q2UBE9 H0ZDT9 A0A182FV47 A0A087V099 A0A341CYT0 A4FV47 A0A2Y9FDE4 A0A384BAJ1 A0A3Q7VW59 H9GZX0 Q3UI99 Q9Z1X9 A0A0A9YQW4 K7GSB7 F1MQQ0 Q5E9W1 A0A1U7TNB8 A0A3Q7U1N8 A0A061I7M6 Q4U3P7 G3QNC9 A0A2R9A6H4 A0A2Y9LBD2 A0A2Y9H4H2 A0A2U3YXH3 O75419 A0A3P4LWF5 A0A2I2Y746 A0A2R9A140 L8I6K2 A0A3Q7MZ45 K7A671 A0A2K5IJU4 B4GTR9 A0A2K6SB67 F7IIN6 A0A2Y9J0S9 A0A2K6P4W2
Pubmed
19121390
26354079
25474469
24845553
28004739
26483478
+ More
28648823 17510324 24508170 25244985 30249741 20798317 21347285 21719571 18362917 19820115 15592404 23371554 20360741 19393038 19892987 10349636 11042159 11076861 11217851 12466851 12040188 16141073 21183079 9724329 10051334 15489334 25401762 26823975 30723633 11282978 16305752 23929341 15753125 22398555 22722832 9755170 9660782 12975309 14702039 15461802 10591208 18669648 21269460 22814378 25933514 27374770 22751099 17994087 25362486
28648823 17510324 24508170 25244985 30249741 20798317 21347285 21719571 18362917 19820115 15592404 23371554 20360741 19393038 19892987 10349636 11042159 11076861 11217851 12466851 12040188 16141073 21183079 9724329 10051334 15489334 25401762 26823975 30723633 11282978 16305752 23929341 15753125 22398555 22722832 9755170 9660782 12975309 14702039 15461802 10591208 18669648 21269460 22814378 25933514 27374770 22751099 17994087 25362486
EMBL
BABH01004178
BABH01004179
BABH01004180
BABH01004181
BABH01004182
BABH01004183
+ More
BABH01004184 BABH01004185 BABH01004186 ODYU01003090 SOQ41403.1 NWSH01000086 PCG79773.1 KQ459582 KPI98939.1 KQ459700 KPJ20356.1 ACPB03009329 GBBI01003715 JAC14997.1 KK852802 KDR16293.1 KK854801 PTY18855.1 GFTR01006955 JAW09471.1 NEVH01005904 PNF38168.1 GEZM01057716 JAV72125.1 GGMS01017183 MBY86386.1 GECU01002162 JAT05545.1 GGMR01005577 MBY18196.1 JXUM01081860 KQ563273 KXJ74145.1 GFXV01004157 MBW15962.1 NNAY01000122 OXU30770.1 GEZM01057717 JAV72124.1 ABLF02024971 CH478641 EAT32693.1 KK107570 EZA48863.1 QOIP01000005 RLU22661.1 KQ435709 KOX80006.1 GL448740 EFN83930.1 CH477475 EAT40319.1 GBYB01006107 JAG75874.1 AXCM01007581 AJWK01022752 AJWK01022753 ABLF02039328 KQ976453 KYM85317.1 KQ435134 KZC14878.1 GGMR01019176 MBY31795.1 ADTU01008602 AJVK01023820 AJVK01023821 GL888206 EGI65200.1 KQ777667 OAD52067.1 KQ971348 EFA05648.1 KQ982708 KYQ51812.1 UFQS01001695 UFQT01001695 SSX11889.1 SSX31451.1 JXJN01012503 GFXV01007061 MBW18866.1 KQ414924 KOC59400.1 AXCN02001081 KZ288442 PBC25700.1 QCYY01001827 ROT74957.1 AADN05000505 KB376592 AKCR02000030 EMC88936.1 PKK25989.1 ABQF01029280 KK122584 KFM83038.1 BC123732 AAI23733.1 AK147013 CH466521 BAE27607.1 EDK97534.1 AF098068 AJ223729 AF081536 AF081537 AF081538 AF081539 BC028635 GBHO01011684 GDHC01021402 JAG31920.1 JAP97226.1 AEMK02000096 DQIR01245662 DQIR01259016 HDC01140.1 HDC14494.1 BT020809 AAX08826.1 KE674677 ERE76198.1 DQ016322 AAY41172.1 CABD030119964 CABD030119965 CABD030119966 CABD030119967 AJFE02045136 AJFE02045137 AF062495 AF053074 AJ223728 AF081535 AY358971 AK293123 AK293338 BT006792 AY572790 CT841513 AC000082 AC000087 AC000088 CH471176 BC006232 BC010022 CYRY02003558 VCW68172.1 JH882094 ELR50902.1 GABC01007453 GABF01008944 GABD01005361 GABE01003596 NBAG03000088 JAA03885.1 JAA13201.1 JAA27739.1 JAA41143.1 PNI90392.1 PNI90396.1 CH479190 EDW25939.1
BABH01004184 BABH01004185 BABH01004186 ODYU01003090 SOQ41403.1 NWSH01000086 PCG79773.1 KQ459582 KPI98939.1 KQ459700 KPJ20356.1 ACPB03009329 GBBI01003715 JAC14997.1 KK852802 KDR16293.1 KK854801 PTY18855.1 GFTR01006955 JAW09471.1 NEVH01005904 PNF38168.1 GEZM01057716 JAV72125.1 GGMS01017183 MBY86386.1 GECU01002162 JAT05545.1 GGMR01005577 MBY18196.1 JXUM01081860 KQ563273 KXJ74145.1 GFXV01004157 MBW15962.1 NNAY01000122 OXU30770.1 GEZM01057717 JAV72124.1 ABLF02024971 CH478641 EAT32693.1 KK107570 EZA48863.1 QOIP01000005 RLU22661.1 KQ435709 KOX80006.1 GL448740 EFN83930.1 CH477475 EAT40319.1 GBYB01006107 JAG75874.1 AXCM01007581 AJWK01022752 AJWK01022753 ABLF02039328 KQ976453 KYM85317.1 KQ435134 KZC14878.1 GGMR01019176 MBY31795.1 ADTU01008602 AJVK01023820 AJVK01023821 GL888206 EGI65200.1 KQ777667 OAD52067.1 KQ971348 EFA05648.1 KQ982708 KYQ51812.1 UFQS01001695 UFQT01001695 SSX11889.1 SSX31451.1 JXJN01012503 GFXV01007061 MBW18866.1 KQ414924 KOC59400.1 AXCN02001081 KZ288442 PBC25700.1 QCYY01001827 ROT74957.1 AADN05000505 KB376592 AKCR02000030 EMC88936.1 PKK25989.1 ABQF01029280 KK122584 KFM83038.1 BC123732 AAI23733.1 AK147013 CH466521 BAE27607.1 EDK97534.1 AF098068 AJ223729 AF081536 AF081537 AF081538 AF081539 BC028635 GBHO01011684 GDHC01021402 JAG31920.1 JAP97226.1 AEMK02000096 DQIR01245662 DQIR01259016 HDC01140.1 HDC14494.1 BT020809 AAX08826.1 KE674677 ERE76198.1 DQ016322 AAY41172.1 CABD030119964 CABD030119965 CABD030119966 CABD030119967 AJFE02045136 AJFE02045137 AF062495 AF053074 AJ223728 AF081535 AY358971 AK293123 AK293338 BT006792 AY572790 CT841513 AC000082 AC000087 AC000088 CH471176 BC006232 BC010022 CYRY02003558 VCW68172.1 JH882094 ELR50902.1 GABC01007453 GABF01008944 GABD01005361 GABE01003596 NBAG03000088 JAA03885.1 JAA13201.1 JAA27739.1 JAA41143.1 PNI90392.1 PNI90396.1 CH479190 EDW25939.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000015103
UP000027135
+ More
UP000235965 UP000192223 UP000069940 UP000249989 UP000215335 UP000007819 UP000008820 UP000053097 UP000076408 UP000279307 UP000053105 UP000008237 UP000075883 UP000092461 UP000078540 UP000076502 UP000005205 UP000092462 UP000007755 UP000005203 UP000007266 UP000075809 UP000091820 UP000092460 UP000092445 UP000053825 UP000075886 UP000242457 UP000092443 UP000283509 UP000000539 UP000245320 UP000053872 UP000007754 UP000069272 UP000054359 UP000252040 UP000009136 UP000248484 UP000261681 UP000286642 UP000002281 UP000000589 UP000008227 UP000189704 UP000030759 UP000001519 UP000240080 UP000248483 UP000248481 UP000245341 UP000005640 UP000286641 UP000233080 UP000008744 UP000233220 UP000008225 UP000248482 UP000233200
UP000235965 UP000192223 UP000069940 UP000249989 UP000215335 UP000007819 UP000008820 UP000053097 UP000076408 UP000279307 UP000053105 UP000008237 UP000075883 UP000092461 UP000078540 UP000076502 UP000005205 UP000092462 UP000007755 UP000005203 UP000007266 UP000075809 UP000091820 UP000092460 UP000092445 UP000053825 UP000075886 UP000242457 UP000092443 UP000283509 UP000000539 UP000245320 UP000053872 UP000007754 UP000069272 UP000054359 UP000252040 UP000009136 UP000248484 UP000261681 UP000286642 UP000002281 UP000000589 UP000008227 UP000189704 UP000030759 UP000001519 UP000240080 UP000248483 UP000248481 UP000245341 UP000005640 UP000286641 UP000233080 UP000008744 UP000233220 UP000008225 UP000248482 UP000233200
Interpro
Gene 3D
ProteinModelPortal
H9JU72
A0A2H1VKQ4
A0A2A4K7M8
A0A194Q1R9
A0A194RVZ2
T1HHG2
+ More
A0A023F0J3 A0A067R9J8 A0A2R7WKB3 A0A224XN74 A0A2J7RBG6 A0A1Y1LJK7 A0A2S2R8N5 A0A1W4X9Z5 A0A1B6K275 A0A2S2NN53 A0A182GR65 A0A2H8TQS0 A0A232FJQ4 A0A1Y1LES8 J9K5U7 Q16EN6 A0A026VYI2 A0A182Y9K9 A0A3L8DQJ9 A0A0M9AAS8 E2BK22 Q170F3 A0A0C9QZG8 A0A182M445 A0A1B0CPX3 J9K7X0 A0A195BKW3 A0A154PU39 A0A2S2PQW2 A0A158P3W2 A0A1B0D3Z1 F4WKV8 A0A310SCM5 A0A087ZR06 D2A4C4 A0A151WV55 A0A336L5R6 A0A1A9WVN8 A0A1B0BDI7 A0A1A9ZKR9 A0A2H8TX98 A0A0L7QLB8 A0A182Q3S6 A0A2A3E1X5 A0A1A9YME8 A0A3R7M7E1 E1BYS7 A0A2U4ALI7 R7VUU8 A0A3Q2UBE9 H0ZDT9 A0A182FV47 A0A087V099 A0A341CYT0 A4FV47 A0A2Y9FDE4 A0A384BAJ1 A0A3Q7VW59 H9GZX0 Q3UI99 Q9Z1X9 A0A0A9YQW4 K7GSB7 F1MQQ0 Q5E9W1 A0A1U7TNB8 A0A3Q7U1N8 A0A061I7M6 Q4U3P7 G3QNC9 A0A2R9A6H4 A0A2Y9LBD2 A0A2Y9H4H2 A0A2U3YXH3 O75419 A0A3P4LWF5 A0A2I2Y746 A0A2R9A140 L8I6K2 A0A3Q7MZ45 K7A671 A0A2K5IJU4 B4GTR9 A0A2K6SB67 F7IIN6 A0A2Y9J0S9 A0A2K6P4W2
A0A023F0J3 A0A067R9J8 A0A2R7WKB3 A0A224XN74 A0A2J7RBG6 A0A1Y1LJK7 A0A2S2R8N5 A0A1W4X9Z5 A0A1B6K275 A0A2S2NN53 A0A182GR65 A0A2H8TQS0 A0A232FJQ4 A0A1Y1LES8 J9K5U7 Q16EN6 A0A026VYI2 A0A182Y9K9 A0A3L8DQJ9 A0A0M9AAS8 E2BK22 Q170F3 A0A0C9QZG8 A0A182M445 A0A1B0CPX3 J9K7X0 A0A195BKW3 A0A154PU39 A0A2S2PQW2 A0A158P3W2 A0A1B0D3Z1 F4WKV8 A0A310SCM5 A0A087ZR06 D2A4C4 A0A151WV55 A0A336L5R6 A0A1A9WVN8 A0A1B0BDI7 A0A1A9ZKR9 A0A2H8TX98 A0A0L7QLB8 A0A182Q3S6 A0A2A3E1X5 A0A1A9YME8 A0A3R7M7E1 E1BYS7 A0A2U4ALI7 R7VUU8 A0A3Q2UBE9 H0ZDT9 A0A182FV47 A0A087V099 A0A341CYT0 A4FV47 A0A2Y9FDE4 A0A384BAJ1 A0A3Q7VW59 H9GZX0 Q3UI99 Q9Z1X9 A0A0A9YQW4 K7GSB7 F1MQQ0 Q5E9W1 A0A1U7TNB8 A0A3Q7U1N8 A0A061I7M6 Q4U3P7 G3QNC9 A0A2R9A6H4 A0A2Y9LBD2 A0A2Y9H4H2 A0A2U3YXH3 O75419 A0A3P4LWF5 A0A2I2Y746 A0A2R9A140 L8I6K2 A0A3Q7MZ45 K7A671 A0A2K5IJU4 B4GTR9 A0A2K6SB67 F7IIN6 A0A2Y9J0S9 A0A2K6P4W2
PDB
5DGO
E-value=6.57939e-46,
Score=465
Ontologies
GO
GO:0006270
GO:0051301
GO:0016021
GO:0003676
GO:0008408
GO:0005656
GO:0000727
GO:0003688
GO:0003682
GO:0031938
GO:1900087
GO:0003697
GO:0043138
GO:1902977
GO:0031261
GO:0031298
GO:0005813
GO:0005634
GO:0005654
GO:0000076
GO:0005737
GO:0006260
GO:0000082
GO:0000083
GO:0006277
GO:0030261
GO:0006508
GO:0015074
GO:0004672
GO:0005524
GO:0006468
GO:0006418
GO:0000049
GO:0016876
PANTHER
Topology
Subcellular location
Nucleus
Cytoplasm
Cytoplasm
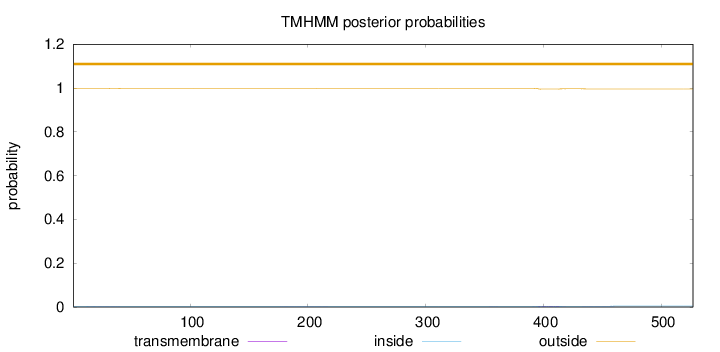
Length:
527
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.13329
Exp number, first 60 AAs:
0.0199
Total prob of N-in:
0.00328
outside
1 - 527
Population Genetic Test Statistics
Pi
168.502424
Theta
146.5509
Tajima's D
0.978797
CLR
1.14748
CSRT
0.647667616619169
Interpretation
Uncertain
