Gene
KWMTBOMO09417
Pre Gene Modal
BGIBMGA012773
Annotation
PREDICTED:_venom_serine_carboxypeptidase-like_[Amyelois_transitella]
Full name
Carboxypeptidase
+ More
Vitellogenic carboxypeptidase
Venom serine carboxypeptidase
Vitellogenic carboxypeptidase
Venom serine carboxypeptidase
Location in the cell
PlasmaMembrane Reliability : 1.014
Sequence
CDS
ATGGCTGTGAAATGGAAGCCTCTGTTGAGCGCAGTGCCGTGTGTTTTGTTTTTTTCTACTTTGTTCCCGCTGAACGGTGCTAAATTTCCGAATGTTTATCCACGAGTGAAGCTGGACGCTCGACCTAATGACTCGGCCGGGGATCCTCTCATGCTGACGCCGTTTCTCAAAAACGAGTCCATAGCCCTCGCCCAGGAACTATCAAGAGTGTCATTGACGGAAACACTTGGGATCGAGAGTCACGCGGGATTCTTTACAGTTGACGAGAAGTTCAATTCAAATATGTACTTTTGGTATTTCCCGCCTTTATCGAAGAACGAAGCTGCACCGGTTTTGCTGTGGCTGCAAGGAGGTCCCGGGGCCAGCTCCTTATTCGGGCTCTTTACGGAAGTCGGGCCGCTGATCGCAAGCAAGGACGGTTTTGCCAAAAGAGAGTATCATTGGGCTAAGAACTATCACCTGATTTTCATCGACAACCCCGTGGGGACCGGCTTCAGTTTCACCGATAAAGATGAAGGTTACTGCACCGATGAGACGTGTATCGCTAAAGGACTCTTCGCAGCAATGGACCAGTTCTTCACTTTGTTCCCGAATTTGAGGGCGAATGATTTCTACATAGCCGGTGAGTCATACGCCGGTAAATACATTCCGGCGTTCGGGTTGTACGTTCACAATCTTAATCAGGAATCGAAGAGAATCAATTTGAAGGGCATGGCCATCGGAAACGGCTACTGCGACCCCCTTCATCAGCTCGACTACGGTGACTATCTGTACCAGCACGGTCTGATAGACGACAACCAACTGAAAACGTTTTTAAAATATCAACTCGAAATTTCGACTCGTATCAAGAAAGGAGAATATTCAACGGCCGCGTATTTGCTCGATGAACTGTTTGATGGTGAAATGACCAAGAATTCAATGTTTAAAACTTATACGGGATTTGAGAATTCATATAATTTTCTGATAAGCAAGGAGGAAGATGATATGACTATTTACGCGGATTTGTTGAAGGATGACACCGTGCGACGCGGCGTGCACGTCGGAGGCCTGGCCTTCAACGACGGCCTGCAAGTGCTGACCAGTTTGGTGGACGATCTGACGCGGTCCGTCGCCCCGCTGCTAGCCAAACTGCTCTCTCACTACCCGATCATGTTCTATAGCGGACAGCTGGACATCATTGTCGCGTACCCTCTGACGGAAAAATTCCTCGCTAACTTGAACTTCTCCGCTGTGGTCGAGTACAAGTTTGCCCCGAGGAACATCTGGAGAGTGGACGGCGACGTCGCTGGTTACGTCAGGAAGGCCGGCAACCTGACCGAGGTGTTGGTTCGGAACGCAGGCCACATGGTGCCGCGGGACCAGCCTAAATGGGCCTTCGAACTAATAACGCAGTTCATCAGTAAAATTTTGTAA
Protein
MAVKWKPLLSAVPCVLFFSTLFPLNGAKFPNVYPRVKLDARPNDSAGDPLMLTPFLKNESIALAQELSRVSLTETLGIESHAGFFTVDEKFNSNMYFWYFPPLSKNEAAPVLLWLQGGPGASSLFGLFTEVGPLIASKDGFAKREYHWAKNYHLIFIDNPVGTGFSFTDKDEGYCTDETCIAKGLFAAMDQFFTLFPNLRANDFYIAGESYAGKYIPAFGLYVHNLNQESKRINLKGMAIGNGYCDPLHQLDYGDYLYQHGLIDDNQLKTFLKYQLEISTRIKKGEYSTAAYLLDELFDGEMTKNSMFKTYTGFENSYNFLISKEEDDMTIYADLLKDDTVRRGVHVGGLAFNDGLQVLTSLVDDLTRSVAPLLAKLLSHYPIMFYSGQLDIIVAYPLTEKFLANLNFSAVVEYKFAPRNIWRVDGDVAGYVRKAGNLTEVLVRNAGHMVPRDQPKWAFELITQFISKIL
Summary
Description
May play a role in activating hydrolytic enzymes that are involved in the degradation of yolk proteins in developing embryos or may function as an exopeptidase in the degradation of vitellogenin.
Catalytic Activity
Release of a C-terminal amino acid with broad specificity.
Similarity
Belongs to the peptidase S10 family.
Keywords
Carboxypeptidase
Complete proteome
Direct protein sequencing
Glycoprotein
Hydrolase
Protease
Reference proteome
Secreted
Signal
Allergen
Feature
chain Carboxypeptidase
Uniprot
H9JTB1
A0A2H1VMC9
A0A2A4K6A6
A0A0L7LM73
A0A194Q6A2
A0A194RSA4
+ More
S4PEN0 A0A2A4K7Y0 A0A212FM71 A0A1B0D870 A0A1B0D2X8 A0A2A4K7Y2 H9JU73 A0A2H1VKQ6 A0A1E1WAZ3 A0A0L7LM79 A0A194RR51 A0A194Q054 A0A1B0CQ02 I4DN75 A0A194RR56 D7EHX7 S4P7T7 A0A2K7P6A8 U4U2S4 P42660 A0A182BVG3 A0A182H8I4 U5EN01 A0A084WNC9 A0A182XYY3 A0A182H8I5 A0A1Q3F9Y2 A0A182VEU0 A0A1Q3F9S6 B0WT67 A0A182X3P0 A0A182I3C9 A0A182MJA6 A0A182U5W4 A0A182WJ64 A0A3R7QLR9 A0A0M8ZQT3 A0A182K218 A0A1Y1LX67 B4LP47 Q7QJG6 A0A182KRE8 Q175U3 A0A182PQD1 A0A194Q1S4 A0A1Q3F8R6 A0A1S4FM29 A0A1W4W9S4 A0A224XJN6 A0A1Q3F8Y0 A0A336M8H4 B0WNR6 B4PNK5 Q16W90 A0A2A3EHH5 B4J5M6 E9IJU5 A0A182J314 A0A182JGG1 A0A182RC12 W5J9B8 A0A1L8DP70 B4LJX3 A0A3B0JNA7 A0A182FF64 A0A182NHT4 A0A170XJP7 Q294S7 B4KNT4 C9WMM5 A0A182QCW5 B4KLQ9 A0A182PVV1 B4NEY8 K7IV06 A0A182K070 A0A182T2C4 B4QST7 A0A1I8MGZ8 A0A182WLV3 A0A1B6EFH6 R4FLC5 A0A182Y0S5 A0A182NMB9 F4X0L6 A0A182QKM9 B4IHR7 A0A182BVG2 A0A0M5J9U2 A0A1J1IBF1 A0A182HBE4 Q9VDT5 A0A195F7F6 A0A182XCU7
S4PEN0 A0A2A4K7Y0 A0A212FM71 A0A1B0D870 A0A1B0D2X8 A0A2A4K7Y2 H9JU73 A0A2H1VKQ6 A0A1E1WAZ3 A0A0L7LM79 A0A194RR51 A0A194Q054 A0A1B0CQ02 I4DN75 A0A194RR56 D7EHX7 S4P7T7 A0A2K7P6A8 U4U2S4 P42660 A0A182BVG3 A0A182H8I4 U5EN01 A0A084WNC9 A0A182XYY3 A0A182H8I5 A0A1Q3F9Y2 A0A182VEU0 A0A1Q3F9S6 B0WT67 A0A182X3P0 A0A182I3C9 A0A182MJA6 A0A182U5W4 A0A182WJ64 A0A3R7QLR9 A0A0M8ZQT3 A0A182K218 A0A1Y1LX67 B4LP47 Q7QJG6 A0A182KRE8 Q175U3 A0A182PQD1 A0A194Q1S4 A0A1Q3F8R6 A0A1S4FM29 A0A1W4W9S4 A0A224XJN6 A0A1Q3F8Y0 A0A336M8H4 B0WNR6 B4PNK5 Q16W90 A0A2A3EHH5 B4J5M6 E9IJU5 A0A182J314 A0A182JGG1 A0A182RC12 W5J9B8 A0A1L8DP70 B4LJX3 A0A3B0JNA7 A0A182FF64 A0A182NHT4 A0A170XJP7 Q294S7 B4KNT4 C9WMM5 A0A182QCW5 B4KLQ9 A0A182PVV1 B4NEY8 K7IV06 A0A182K070 A0A182T2C4 B4QST7 A0A1I8MGZ8 A0A182WLV3 A0A1B6EFH6 R4FLC5 A0A182Y0S5 A0A182NMB9 F4X0L6 A0A182QKM9 B4IHR7 A0A182BVG2 A0A0M5J9U2 A0A1J1IBF1 A0A182HBE4 Q9VDT5 A0A195F7F6 A0A182XCU7
EC Number
3.4.16.-
3.4.16.5
3.4.16.5
Pubmed
19121390
26227816
26354079
23622113
22118469
22651552
+ More
18362917 19820115 23537049 1961751 9087558 17510324 26483478 24438588 25244985 28004739 17994087 12364791 14747013 17210077 20966253 17550304 21282665 20920257 23761445 15632085 17073008 20075255 25315136 21719571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
18362917 19820115 23537049 1961751 9087558 17510324 26483478 24438588 25244985 28004739 17994087 12364791 14747013 17210077 20966253 17550304 21282665 20920257 23761445 15632085 17073008 20075255 25315136 21719571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
BABH01004187
ODYU01003090
SOQ41404.1
NWSH01000086
PCG79771.1
JTDY01000595
+ More
KOB76530.1 KQ459582 KPI98940.1 KQ459700 KPJ20357.1 GAIX01004417 JAA88143.1 PCG79772.1 AGBW02007654 OWR54826.1 AJVK01027239 AJVK01023133 PCG79770.1 SOQ41405.1 GDQN01006939 JAT84115.1 KOB76535.1 KPJ20358.1 KPI98941.1 AJWK01022899 AK402751 BAM19365.1 KPJ20363.1 KQ971323 KYB28573.1 GAIX01004449 JAA88111.1 KB631843 ERL86633.1 M79452 L46594 CH477395 KU179863 AND61388.1 JXUM01029016 KQ560839 KXJ80663.1 GANO01000788 JAB59083.1 ATLV01024587 KE525352 KFB51723.1 KXJ80664.1 GFDL01010661 JAV24384.1 GFDL01010711 JAV24334.1 DS232080 EDS34210.1 APCN01000205 AXCM01000691 QCYY01000750 ROT83024.1 KQ435896 KOX69240.1 GEZM01050301 JAV75627.1 CH940648 EDW62241.1 AAAB01008807 EAA04657.4 EAT41862.1 KPI98944.1 GFDL01011075 JAV23970.1 GFTR01006388 JAW10038.1 GFDL01011072 JAV23973.1 UFQT01000606 SSX25661.1 DS232014 EDS31907.1 CM000160 EDW96068.1 CH477572 EAT38860.1 KZ288242 PBC31235.1 CH916367 EDW01802.1 GL763883 EFZ19168.1 ADMH02001806 ETN61052.1 GFDF01005801 JAV08283.1 EDW61627.1 OUUW01000008 SPP83704.1 GEMB01004337 JAR98936.1 CM000070 EAL28887.1 CH933808 EDW08979.1 FJ765738 AXCN02000935 EDW08695.1 CH964251 EDW83363.1 AAZX01001162 CM000364 EDX12297.1 GEDC01000657 JAS36641.1 ACPB03001109 GAHY01001473 JAA76037.1 GL888498 EGI60008.1 AXCN02000377 CH480840 EDW49434.1 KU179862 AND61387.1 CP012524 ALC40556.1 CVRI01000047 CRK97539.1 JXUM01031871 KQ560938 KXJ80263.1 AE014297 AY052022 BT099565 AAF55705.1 AAK93446.1 ACU33957.1 KQ981744 KYN36373.1
KOB76530.1 KQ459582 KPI98940.1 KQ459700 KPJ20357.1 GAIX01004417 JAA88143.1 PCG79772.1 AGBW02007654 OWR54826.1 AJVK01027239 AJVK01023133 PCG79770.1 SOQ41405.1 GDQN01006939 JAT84115.1 KOB76535.1 KPJ20358.1 KPI98941.1 AJWK01022899 AK402751 BAM19365.1 KPJ20363.1 KQ971323 KYB28573.1 GAIX01004449 JAA88111.1 KB631843 ERL86633.1 M79452 L46594 CH477395 KU179863 AND61388.1 JXUM01029016 KQ560839 KXJ80663.1 GANO01000788 JAB59083.1 ATLV01024587 KE525352 KFB51723.1 KXJ80664.1 GFDL01010661 JAV24384.1 GFDL01010711 JAV24334.1 DS232080 EDS34210.1 APCN01000205 AXCM01000691 QCYY01000750 ROT83024.1 KQ435896 KOX69240.1 GEZM01050301 JAV75627.1 CH940648 EDW62241.1 AAAB01008807 EAA04657.4 EAT41862.1 KPI98944.1 GFDL01011075 JAV23970.1 GFTR01006388 JAW10038.1 GFDL01011072 JAV23973.1 UFQT01000606 SSX25661.1 DS232014 EDS31907.1 CM000160 EDW96068.1 CH477572 EAT38860.1 KZ288242 PBC31235.1 CH916367 EDW01802.1 GL763883 EFZ19168.1 ADMH02001806 ETN61052.1 GFDF01005801 JAV08283.1 EDW61627.1 OUUW01000008 SPP83704.1 GEMB01004337 JAR98936.1 CM000070 EAL28887.1 CH933808 EDW08979.1 FJ765738 AXCN02000935 EDW08695.1 CH964251 EDW83363.1 AAZX01001162 CM000364 EDX12297.1 GEDC01000657 JAS36641.1 ACPB03001109 GAHY01001473 JAA76037.1 GL888498 EGI60008.1 AXCN02000377 CH480840 EDW49434.1 KU179862 AND61387.1 CP012524 ALC40556.1 CVRI01000047 CRK97539.1 JXUM01031871 KQ560938 KXJ80263.1 AE014297 AY052022 BT099565 AAF55705.1 AAK93446.1 ACU33957.1 KQ981744 KYN36373.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053268
UP000053240
UP000007151
+ More
UP000092462 UP000092461 UP000007266 UP000030742 UP000008820 UP000069940 UP000249989 UP000030765 UP000076408 UP000075903 UP000002320 UP000076407 UP000075840 UP000075883 UP000075902 UP000075920 UP000283509 UP000053105 UP000075881 UP000008792 UP000007062 UP000075882 UP000075885 UP000192221 UP000002282 UP000242457 UP000001070 UP000075880 UP000075900 UP000000673 UP000268350 UP000069272 UP000075884 UP000001819 UP000009192 UP000005203 UP000075886 UP000007798 UP000002358 UP000075901 UP000000304 UP000095301 UP000015103 UP000007755 UP000001292 UP000092553 UP000183832 UP000000803 UP000078541
UP000092462 UP000092461 UP000007266 UP000030742 UP000008820 UP000069940 UP000249989 UP000030765 UP000076408 UP000075903 UP000002320 UP000076407 UP000075840 UP000075883 UP000075902 UP000075920 UP000283509 UP000053105 UP000075881 UP000008792 UP000007062 UP000075882 UP000075885 UP000192221 UP000002282 UP000242457 UP000001070 UP000075880 UP000075900 UP000000673 UP000268350 UP000069272 UP000075884 UP000001819 UP000009192 UP000005203 UP000075886 UP000007798 UP000002358 UP000075901 UP000000304 UP000095301 UP000015103 UP000007755 UP000001292 UP000092553 UP000183832 UP000000803 UP000078541
Pfam
PF00450 Peptidase_S10
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
H9JTB1
A0A2H1VMC9
A0A2A4K6A6
A0A0L7LM73
A0A194Q6A2
A0A194RSA4
+ More
S4PEN0 A0A2A4K7Y0 A0A212FM71 A0A1B0D870 A0A1B0D2X8 A0A2A4K7Y2 H9JU73 A0A2H1VKQ6 A0A1E1WAZ3 A0A0L7LM79 A0A194RR51 A0A194Q054 A0A1B0CQ02 I4DN75 A0A194RR56 D7EHX7 S4P7T7 A0A2K7P6A8 U4U2S4 P42660 A0A182BVG3 A0A182H8I4 U5EN01 A0A084WNC9 A0A182XYY3 A0A182H8I5 A0A1Q3F9Y2 A0A182VEU0 A0A1Q3F9S6 B0WT67 A0A182X3P0 A0A182I3C9 A0A182MJA6 A0A182U5W4 A0A182WJ64 A0A3R7QLR9 A0A0M8ZQT3 A0A182K218 A0A1Y1LX67 B4LP47 Q7QJG6 A0A182KRE8 Q175U3 A0A182PQD1 A0A194Q1S4 A0A1Q3F8R6 A0A1S4FM29 A0A1W4W9S4 A0A224XJN6 A0A1Q3F8Y0 A0A336M8H4 B0WNR6 B4PNK5 Q16W90 A0A2A3EHH5 B4J5M6 E9IJU5 A0A182J314 A0A182JGG1 A0A182RC12 W5J9B8 A0A1L8DP70 B4LJX3 A0A3B0JNA7 A0A182FF64 A0A182NHT4 A0A170XJP7 Q294S7 B4KNT4 C9WMM5 A0A182QCW5 B4KLQ9 A0A182PVV1 B4NEY8 K7IV06 A0A182K070 A0A182T2C4 B4QST7 A0A1I8MGZ8 A0A182WLV3 A0A1B6EFH6 R4FLC5 A0A182Y0S5 A0A182NMB9 F4X0L6 A0A182QKM9 B4IHR7 A0A182BVG2 A0A0M5J9U2 A0A1J1IBF1 A0A182HBE4 Q9VDT5 A0A195F7F6 A0A182XCU7
S4PEN0 A0A2A4K7Y0 A0A212FM71 A0A1B0D870 A0A1B0D2X8 A0A2A4K7Y2 H9JU73 A0A2H1VKQ6 A0A1E1WAZ3 A0A0L7LM79 A0A194RR51 A0A194Q054 A0A1B0CQ02 I4DN75 A0A194RR56 D7EHX7 S4P7T7 A0A2K7P6A8 U4U2S4 P42660 A0A182BVG3 A0A182H8I4 U5EN01 A0A084WNC9 A0A182XYY3 A0A182H8I5 A0A1Q3F9Y2 A0A182VEU0 A0A1Q3F9S6 B0WT67 A0A182X3P0 A0A182I3C9 A0A182MJA6 A0A182U5W4 A0A182WJ64 A0A3R7QLR9 A0A0M8ZQT3 A0A182K218 A0A1Y1LX67 B4LP47 Q7QJG6 A0A182KRE8 Q175U3 A0A182PQD1 A0A194Q1S4 A0A1Q3F8R6 A0A1S4FM29 A0A1W4W9S4 A0A224XJN6 A0A1Q3F8Y0 A0A336M8H4 B0WNR6 B4PNK5 Q16W90 A0A2A3EHH5 B4J5M6 E9IJU5 A0A182J314 A0A182JGG1 A0A182RC12 W5J9B8 A0A1L8DP70 B4LJX3 A0A3B0JNA7 A0A182FF64 A0A182NHT4 A0A170XJP7 Q294S7 B4KNT4 C9WMM5 A0A182QCW5 B4KLQ9 A0A182PVV1 B4NEY8 K7IV06 A0A182K070 A0A182T2C4 B4QST7 A0A1I8MGZ8 A0A182WLV3 A0A1B6EFH6 R4FLC5 A0A182Y0S5 A0A182NMB9 F4X0L6 A0A182QKM9 B4IHR7 A0A182BVG2 A0A0M5J9U2 A0A1J1IBF1 A0A182HBE4 Q9VDT5 A0A195F7F6 A0A182XCU7
PDB
4MWT
E-value=1.08325e-36,
Score=385
Ontologies
GO
PANTHER
Topology
Subcellular location
Secreted
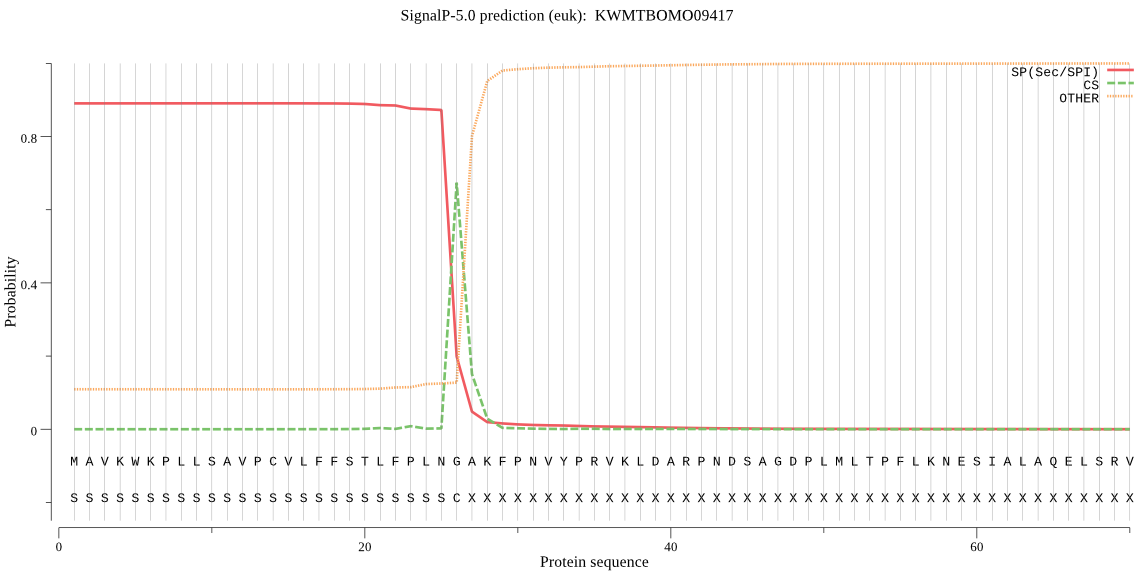
SignalP
Position: 1 - 26,
Likelihood: 0.890541
Length:
470
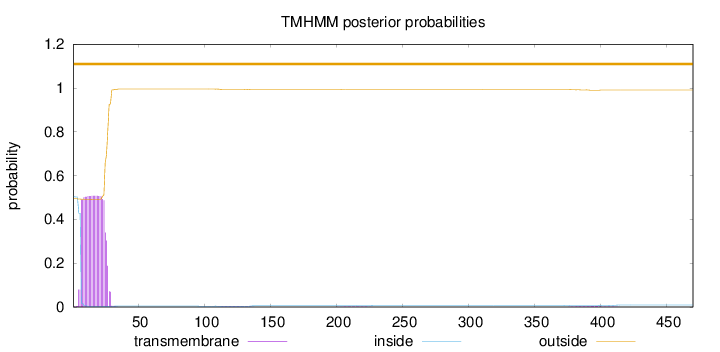
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
10.42754
Exp number, first 60 AAs:
10.20153
Total prob of N-in:
0.50605
POSSIBLE N-term signal
sequence
outside
1 - 470
Population Genetic Test Statistics
Pi
180.165997
Theta
41.911715
Tajima's D
0.380247
CLR
0
CSRT
0.48107594620269
Interpretation
Uncertain
