Gene
KWMTBOMO09415 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012772
Annotation
PREDICTED:_ATP-dependent_RNA_helicase_WM6_[Amyelois_transitella]
Full name
ATP-dependent RNA helicase WM6
Alternative Name
HEL/UAP56
Location in the cell
Nuclear Reliability : 2.74
Sequence
CDS
ATGGCTGACAACGACGATCTTCTCGACTACGAAGATGAAGAACAAGCAGATCAACAGACTGCTGACGGGTCTACCGAAGTGGCGCCAAAGAAGGAGGTTAAAGGATCCTACGTATCCATCCACAGTTCTGGTTTTAGAGACTTCTTACTTAAGCCAGAGATACTCCGCGCTATTGTTGATTGCGGTTTTGAGCATCCTTCCGAAGTTCAACACGAATGCATTCCGCAAGCCGTCCTCGGTATGGACATACTTTGCCAAGCCAAGTCCGGTATGGGTAAGACAGCCGTGTTTGTTTTGGCAACATTGCAGCAGCTGGAACCCTCCGAGAGCCACGTTTACGTGCTTGTAATGTGCCACACCAGAGAGTTGGCTTTCCAAATAAGTAAGGAGTACGAGCGTTTCTCGAAATATATGTCAGGAGTCAGAGTATCAGTATTCTTTGGTGGTATGCCAATTCAGAAAGACGAGGAAGTCCTTAAGACAGCATGTCCTCACATTGTAGTGGGAACTCCCGGTAGAATTTTGGCATTAGTAAACAGTAAGAAACTTAATTTGAAACATTTAAAACACTTCATCCTTGATGAGTGTGATAAGATGCTGGAATCTCTAGACATGAGGCGAGATGTGCAGGAGATATTCCGGAACACTCCTCACGGGAAACAGGTGATGATGTTCTCAGCCACATTGAGTAAAGAAATCAGACCTGTGTGTAAGAAATTTATGCAAGACCCAATGGAAGTTTATGTAGATGATGAAGCCAAACTCAAGCTACATGGACTGCAGCAACATTATGTGAAACTAAAAGAAAATGAGAAGAATAAAAAACTCTTCGAACTACTTGATGTATTGGAGTTCAATCAGGTGGTTATTTTTGTGAAATCAGTACAGCGCTGCATAGCTCTGGCACAACTGCTCACCGACCAAAACTTCCCTGCCATTGGTATCCACAGAAATATGACCCAGGATGAGCGTCTCTCCCGCTATCAACAGTTCAAGGACTTCCAAAAAAGGATCCTGGTTGCAACAAATTTGTTTGGCCGGGGAATGGACATTGAAAGGGTAAATATTGTCTTCAACTATGACATGCCTGAAGATTCAGATACATATCTACATCGTGTCGCCCGCGCAGGAAGGTTTGGTACCAAAGGTCTGGCCATTACTATGATATCTGATGAAAATGATGCCAAGATCTTAAACCAAGTACAAGATCGTTTCGATGTGAACATTACAGAGCTGCCTGAAGAAATTGAGCTCTCTACCTACATCGAAGGACGATAG
Protein
MADNDDLLDYEDEEQADQQTADGSTEVAPKKEVKGSYVSIHSSGFRDFLLKPEILRAIVDCGFEHPSEVQHECIPQAVLGMDILCQAKSGMGKTAVFVLATLQQLEPSESHVYVLVMCHTRELAFQISKEYERFSKYMSGVRVSVFFGGMPIQKDEEVLKTACPHIVVGTPGRILALVNSKKLNLKHLKHFILDECDKMLESLDMRRDVQEIFRNTPHGKQVMMFSATLSKEIRPVCKKFMQDPMEVYVDDEAKLKLHGLQQHYVKLKENEKNKKLFELLDVLEFNQVVIFVKSVQRCIALAQLLTDQNFPAIGIHRNMTQDERLSRYQQFKDFQKRILVATNLFGRGMDIERVNIVFNYDMPEDSDTYLHRVARAGRFGTKGLAITMISDENDAKILNQVQDRFDVNITELPEEIELSTYIEGR
Summary
Description
Required for mRNA export out of the nucleus. Probable RNA helicase that may regulate entry into mitosis by down-regulating the expression of other genes whose activity may be rate-limiting for entry into mitosis during embryogenesis. Binds to salivary gland chromosomes and modifies position effect variegation. Promotes an open chromatin structure that favors transcription during development by regulating the spread of heterochromatin.
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Subunit
Component of the spliceosome (Probable). Interacts with the exon junction complex.
Similarity
Belongs to the DEAD box helicase family. DECD subfamily.
Belongs to the glycosyl hydrolase 18 family.
Belongs to the glycosyl hydrolase 18 family.
Keywords
ATP-binding
Complete proteome
Helicase
Hydrolase
mRNA processing
mRNA splicing
Nucleotide-binding
Nucleus
Reference proteome
RNA-binding
Spliceosome
Feature
chain ATP-dependent RNA helicase WM6
Uniprot
H9JTB0
S4NNL7
A0A2A4K6S1
A0A2H1VKS8
A0A194RSI8
I4DJM2
+ More
A0A212FM47 A0A1L8DRS1 K7J0K7 A0A2J7RMJ1 A0A067RV07 V9IIX6 A0A411G6H1 T1PBI2 B4LU13 B4KF36 A0A0M5J9L8 Q1HR75 B4P055 A0A158NA44 B3N4X2 A0A026VV72 A0A1Q3EYG3 U5EWP6 A0A1I8PD82 B0X3U8 W8BPM9 A0A0K8TZP5 A0A034WST2 Q8MYL5 A0A0A1XR85 A0A336M0V1 A0A1W4UXM1 T1EA73 A0A1J1IN60 A0A1Y1M1K1 A0A182MDS0 A0A182R6H4 B4I1H2 B4NRX8 Q27268 D6WLU7 A0A182WH84 E2AH52 A0A0P6IZC8 A0A2M3Z1P2 A0A2M4BPB1 W5JTU8 A0A154PNY9 A0A1A9X652 A0A1A9VTL9 A0A1B0AS58 B3MMZ5 B4JAF3 A0A1B0A5V8 J3JZ95 A0A1W4WKZ6 A0A1A9X0P8 A0A182V770 A0A182YFG3 A0A1S4H010 B4MWD1 A0A182I4W6 A0A182PDF8 A0A182JUC4 A0A182UA76 A0A182X2Y9 A0A182R9S6 V5I9W2 A0A0N0BED2 A0A0L0C8Z1 K7J0K9 A0A182M9L9 A0A182QFS2 A0A182NF91 A0A084VY69 Q29NU1 A0A195CV96 E9IXM7 A2I858 A0A195DLS0 F4X1F1 A0A3B0KAV0 A0A195BKD3 A0A336LYS3 A0A2D1QUG1 A0A182KUT1 A0A1B6CLH7 A0A1D2MQE0 A0A1B6HLK6 A0A0A9YR59 A0A1B0FEQ7 A0A0L7RFR5 D3PGA2 A0A384DT24 A0A423TBN8 A0A452TBZ9 G1M447 A0A384A4F9 G1PDC3
A0A212FM47 A0A1L8DRS1 K7J0K7 A0A2J7RMJ1 A0A067RV07 V9IIX6 A0A411G6H1 T1PBI2 B4LU13 B4KF36 A0A0M5J9L8 Q1HR75 B4P055 A0A158NA44 B3N4X2 A0A026VV72 A0A1Q3EYG3 U5EWP6 A0A1I8PD82 B0X3U8 W8BPM9 A0A0K8TZP5 A0A034WST2 Q8MYL5 A0A0A1XR85 A0A336M0V1 A0A1W4UXM1 T1EA73 A0A1J1IN60 A0A1Y1M1K1 A0A182MDS0 A0A182R6H4 B4I1H2 B4NRX8 Q27268 D6WLU7 A0A182WH84 E2AH52 A0A0P6IZC8 A0A2M3Z1P2 A0A2M4BPB1 W5JTU8 A0A154PNY9 A0A1A9X652 A0A1A9VTL9 A0A1B0AS58 B3MMZ5 B4JAF3 A0A1B0A5V8 J3JZ95 A0A1W4WKZ6 A0A1A9X0P8 A0A182V770 A0A182YFG3 A0A1S4H010 B4MWD1 A0A182I4W6 A0A182PDF8 A0A182JUC4 A0A182UA76 A0A182X2Y9 A0A182R9S6 V5I9W2 A0A0N0BED2 A0A0L0C8Z1 K7J0K9 A0A182M9L9 A0A182QFS2 A0A182NF91 A0A084VY69 Q29NU1 A0A195CV96 E9IXM7 A2I858 A0A195DLS0 F4X1F1 A0A3B0KAV0 A0A195BKD3 A0A336LYS3 A0A2D1QUG1 A0A182KUT1 A0A1B6CLH7 A0A1D2MQE0 A0A1B6HLK6 A0A0A9YR59 A0A1B0FEQ7 A0A0L7RFR5 D3PGA2 A0A384DT24 A0A423TBN8 A0A452TBZ9 G1M447 A0A384A4F9 G1PDC3
EC Number
3.6.4.13
Pubmed
19121390
23622113
26354079
22651552
22118469
20075255
+ More
24845553 25315136 17994087 17204158 17510324 17550304 21347285 24508170 30249741 24495485 25348373 12015125 25830018 28004739 22936249 7808417 9215899 10731132 12537572 12537569 11696332 12743041 18362917 19820115 20798317 20920257 23761445 22516182 23537049 25244985 12364791 26108605 24438588 15632085 21282665 17519023 21719571 20966253 27289101 25401762 24813606 20010809 21993624
24845553 25315136 17994087 17204158 17510324 17550304 21347285 24508170 30249741 24495485 25348373 12015125 25830018 28004739 22936249 7808417 9215899 10731132 12537572 12537569 11696332 12743041 18362917 19820115 20798317 20920257 23761445 22516182 23537049 25244985 12364791 26108605 24438588 15632085 21282665 17519023 21719571 20966253 27289101 25401762 24813606 20010809 21993624
EMBL
BABH01004187
GAIX01013881
JAA78679.1
NWSH01000086
PCG79769.1
ODYU01003090
+ More
SOQ41406.1 KQ459700 KPJ20359.1 AK401490 KQ459582 BAM18112.1 KPI98942.1 AGBW02007654 OWR54828.1 GFDF01004908 JAV09176.1 NEVH01002553 PNF42055.1 KK852451 KDR23694.1 JR047589 AEY60451.1 MH365681 QBB01490.1 KA646126 AFP60755.1 CH940649 EDW65066.1 CH933807 EDW13019.1 CP012523 ALC40118.1 DQ440219 CH477210 ABF18252.1 EAT47701.1 CM000157 EDW88920.1 ADTU01009963 CH954177 EDV57874.1 KK107796 QOIP01000013 EZA47627.1 RLU15501.1 GFDL01014702 JAV20343.1 GANO01001323 JAB58548.1 DS232322 EDS40025.1 GAMC01003400 JAC03156.1 GDHF01032350 JAI19964.1 GAKP01001732 JAC57220.1 AJ428513 CAD21558.1 GBXI01000902 JAD13390.1 UFQT01000295 SSX22951.1 GAMD01000687 JAB00904.1 CVRI01000055 CRL01600.1 GEZM01044472 JAV78195.1 AXCM01003231 CH480820 EDW54379.1 CH981625 CM002910 EDX15356.1 KMY88313.1 X79802 L06018 AE014134 AY118921 KQ971343 EFA04166.1 GL439483 EFN67169.1 GDIQ01002134 JAN92603.1 GGFM01001701 MBW22452.1 GGFJ01005640 MBW54781.1 ADMH02000504 ETN66174.1 KQ435007 KZC13579.1 JXJN01002673 CH902620 EDV31020.1 CH916368 EDW03824.1 APGK01051866 BT128576 KB741207 KB632327 AEE63533.1 ENN72901.1 ERL92518.1 AAAB01008984 CH963857 EDW76001.1 APCN01003737 GALX01002240 JAB66226.1 KQ435830 KOX71742.1 JRES01000736 KNC28883.1 AXCM01001875 AXCN02000824 ATLV01018323 KE525231 KFB42913.1 CH379058 EAL34553.2 KQ977276 KYN04462.1 GL766762 EFZ14680.1 EF173367 ABM68586.1 KQ980734 KYN13791.1 GL888529 EGI59746.1 OUUW01000006 SPP82171.1 KQ976455 KYM85108.1 SSX22950.1 KY285064 ATP16167.1 GEDC01022971 GEDC01003436 JAS14327.1 JAS33862.1 LJIJ01000685 ODM95320.1 GECU01032251 JAS75455.1 GBHO01008920 GBRD01011920 JAG34684.1 JAG53904.1 CCAG010014746 KQ414598 KOC69827.1 BT120658 BT121851 HACA01004213 ADD24298.1 ADD38781.1 CDW21574.1 QCYY01001978 ROT73866.1 ACTA01049948 AAPE02040635
SOQ41406.1 KQ459700 KPJ20359.1 AK401490 KQ459582 BAM18112.1 KPI98942.1 AGBW02007654 OWR54828.1 GFDF01004908 JAV09176.1 NEVH01002553 PNF42055.1 KK852451 KDR23694.1 JR047589 AEY60451.1 MH365681 QBB01490.1 KA646126 AFP60755.1 CH940649 EDW65066.1 CH933807 EDW13019.1 CP012523 ALC40118.1 DQ440219 CH477210 ABF18252.1 EAT47701.1 CM000157 EDW88920.1 ADTU01009963 CH954177 EDV57874.1 KK107796 QOIP01000013 EZA47627.1 RLU15501.1 GFDL01014702 JAV20343.1 GANO01001323 JAB58548.1 DS232322 EDS40025.1 GAMC01003400 JAC03156.1 GDHF01032350 JAI19964.1 GAKP01001732 JAC57220.1 AJ428513 CAD21558.1 GBXI01000902 JAD13390.1 UFQT01000295 SSX22951.1 GAMD01000687 JAB00904.1 CVRI01000055 CRL01600.1 GEZM01044472 JAV78195.1 AXCM01003231 CH480820 EDW54379.1 CH981625 CM002910 EDX15356.1 KMY88313.1 X79802 L06018 AE014134 AY118921 KQ971343 EFA04166.1 GL439483 EFN67169.1 GDIQ01002134 JAN92603.1 GGFM01001701 MBW22452.1 GGFJ01005640 MBW54781.1 ADMH02000504 ETN66174.1 KQ435007 KZC13579.1 JXJN01002673 CH902620 EDV31020.1 CH916368 EDW03824.1 APGK01051866 BT128576 KB741207 KB632327 AEE63533.1 ENN72901.1 ERL92518.1 AAAB01008984 CH963857 EDW76001.1 APCN01003737 GALX01002240 JAB66226.1 KQ435830 KOX71742.1 JRES01000736 KNC28883.1 AXCM01001875 AXCN02000824 ATLV01018323 KE525231 KFB42913.1 CH379058 EAL34553.2 KQ977276 KYN04462.1 GL766762 EFZ14680.1 EF173367 ABM68586.1 KQ980734 KYN13791.1 GL888529 EGI59746.1 OUUW01000006 SPP82171.1 KQ976455 KYM85108.1 SSX22950.1 KY285064 ATP16167.1 GEDC01022971 GEDC01003436 JAS14327.1 JAS33862.1 LJIJ01000685 ODM95320.1 GECU01032251 JAS75455.1 GBHO01008920 GBRD01011920 JAG34684.1 JAG53904.1 CCAG010014746 KQ414598 KOC69827.1 BT120658 BT121851 HACA01004213 ADD24298.1 ADD38781.1 CDW21574.1 QCYY01001978 ROT73866.1 ACTA01049948 AAPE02040635
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000002358
+ More
UP000235965 UP000027135 UP000095301 UP000008792 UP000009192 UP000092553 UP000008820 UP000002282 UP000005205 UP000008711 UP000053097 UP000279307 UP000095300 UP000002320 UP000192221 UP000183832 UP000075883 UP000075900 UP000001292 UP000000304 UP000000803 UP000007266 UP000075920 UP000000311 UP000000673 UP000076502 UP000092443 UP000078200 UP000092460 UP000007801 UP000001070 UP000092445 UP000019118 UP000030742 UP000192223 UP000091820 UP000075903 UP000076408 UP000007798 UP000075840 UP000075885 UP000075881 UP000075902 UP000076407 UP000053105 UP000037069 UP000075886 UP000075884 UP000030765 UP000001819 UP000078542 UP000078492 UP000007755 UP000268350 UP000078540 UP000075882 UP000094527 UP000092444 UP000053825 UP000261680 UP000291021 UP000283509 UP000008912 UP000261681 UP000001074
UP000235965 UP000027135 UP000095301 UP000008792 UP000009192 UP000092553 UP000008820 UP000002282 UP000005205 UP000008711 UP000053097 UP000279307 UP000095300 UP000002320 UP000192221 UP000183832 UP000075883 UP000075900 UP000001292 UP000000304 UP000000803 UP000007266 UP000075920 UP000000311 UP000000673 UP000076502 UP000092443 UP000078200 UP000092460 UP000007801 UP000001070 UP000092445 UP000019118 UP000030742 UP000192223 UP000091820 UP000075903 UP000076408 UP000007798 UP000075840 UP000075885 UP000075881 UP000075902 UP000076407 UP000053105 UP000037069 UP000075886 UP000075884 UP000030765 UP000001819 UP000078542 UP000078492 UP000007755 UP000268350 UP000078540 UP000075882 UP000094527 UP000092444 UP000053825 UP000261680 UP000291021 UP000283509 UP000008912 UP000261681 UP000001074
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JTB0
S4NNL7
A0A2A4K6S1
A0A2H1VKS8
A0A194RSI8
I4DJM2
+ More
A0A212FM47 A0A1L8DRS1 K7J0K7 A0A2J7RMJ1 A0A067RV07 V9IIX6 A0A411G6H1 T1PBI2 B4LU13 B4KF36 A0A0M5J9L8 Q1HR75 B4P055 A0A158NA44 B3N4X2 A0A026VV72 A0A1Q3EYG3 U5EWP6 A0A1I8PD82 B0X3U8 W8BPM9 A0A0K8TZP5 A0A034WST2 Q8MYL5 A0A0A1XR85 A0A336M0V1 A0A1W4UXM1 T1EA73 A0A1J1IN60 A0A1Y1M1K1 A0A182MDS0 A0A182R6H4 B4I1H2 B4NRX8 Q27268 D6WLU7 A0A182WH84 E2AH52 A0A0P6IZC8 A0A2M3Z1P2 A0A2M4BPB1 W5JTU8 A0A154PNY9 A0A1A9X652 A0A1A9VTL9 A0A1B0AS58 B3MMZ5 B4JAF3 A0A1B0A5V8 J3JZ95 A0A1W4WKZ6 A0A1A9X0P8 A0A182V770 A0A182YFG3 A0A1S4H010 B4MWD1 A0A182I4W6 A0A182PDF8 A0A182JUC4 A0A182UA76 A0A182X2Y9 A0A182R9S6 V5I9W2 A0A0N0BED2 A0A0L0C8Z1 K7J0K9 A0A182M9L9 A0A182QFS2 A0A182NF91 A0A084VY69 Q29NU1 A0A195CV96 E9IXM7 A2I858 A0A195DLS0 F4X1F1 A0A3B0KAV0 A0A195BKD3 A0A336LYS3 A0A2D1QUG1 A0A182KUT1 A0A1B6CLH7 A0A1D2MQE0 A0A1B6HLK6 A0A0A9YR59 A0A1B0FEQ7 A0A0L7RFR5 D3PGA2 A0A384DT24 A0A423TBN8 A0A452TBZ9 G1M447 A0A384A4F9 G1PDC3
A0A212FM47 A0A1L8DRS1 K7J0K7 A0A2J7RMJ1 A0A067RV07 V9IIX6 A0A411G6H1 T1PBI2 B4LU13 B4KF36 A0A0M5J9L8 Q1HR75 B4P055 A0A158NA44 B3N4X2 A0A026VV72 A0A1Q3EYG3 U5EWP6 A0A1I8PD82 B0X3U8 W8BPM9 A0A0K8TZP5 A0A034WST2 Q8MYL5 A0A0A1XR85 A0A336M0V1 A0A1W4UXM1 T1EA73 A0A1J1IN60 A0A1Y1M1K1 A0A182MDS0 A0A182R6H4 B4I1H2 B4NRX8 Q27268 D6WLU7 A0A182WH84 E2AH52 A0A0P6IZC8 A0A2M3Z1P2 A0A2M4BPB1 W5JTU8 A0A154PNY9 A0A1A9X652 A0A1A9VTL9 A0A1B0AS58 B3MMZ5 B4JAF3 A0A1B0A5V8 J3JZ95 A0A1W4WKZ6 A0A1A9X0P8 A0A182V770 A0A182YFG3 A0A1S4H010 B4MWD1 A0A182I4W6 A0A182PDF8 A0A182JUC4 A0A182UA76 A0A182X2Y9 A0A182R9S6 V5I9W2 A0A0N0BED2 A0A0L0C8Z1 K7J0K9 A0A182M9L9 A0A182QFS2 A0A182NF91 A0A084VY69 Q29NU1 A0A195CV96 E9IXM7 A2I858 A0A195DLS0 F4X1F1 A0A3B0KAV0 A0A195BKD3 A0A336LYS3 A0A2D1QUG1 A0A182KUT1 A0A1B6CLH7 A0A1D2MQE0 A0A1B6HLK6 A0A0A9YR59 A0A1B0FEQ7 A0A0L7RFR5 D3PGA2 A0A384DT24 A0A423TBN8 A0A452TBZ9 G1M447 A0A384A4F9 G1PDC3
PDB
1XTI
E-value=5.6586e-180,
Score=1620
Ontologies
PATHWAY
GO
Topology
Subcellular location
Nucleus speckle
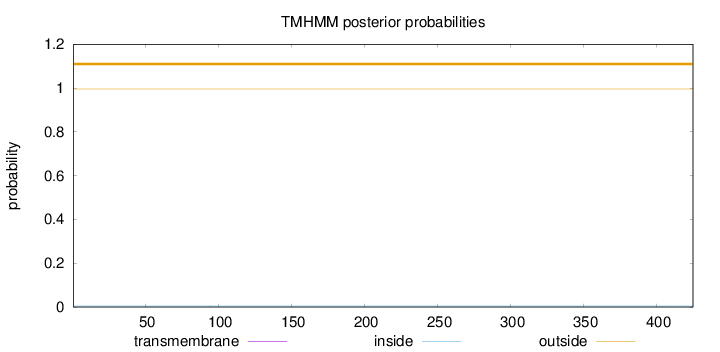
Length:
425
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00249
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00492
outside
1 - 425
Population Genetic Test Statistics
Pi
146.851338
Theta
130.412303
Tajima's D
-0.651738
CLR
1.301359
CSRT
0.205689715514224
Interpretation
Uncertain
