Gene
KWMTBOMO09413
Pre Gene Modal
BGIBMGA013086
Annotation
PREDICTED:_crossover_junction_endonuclease_EME1_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 1.021 Nuclear Reliability : 1.065
Sequence
CDS
ATGAATAAGATTTATAAGCCAGGAGAATGTATGAAGTATATGTACGTGGAGATTCATCCTGTGCTAGCTGAAGCTTGGTACATGGCGGACGTCTCACGGGAGGTGACTTCTGCGTCTGCCCATCTCATAACCACATCAAGTATCTGCGACCCTGCTTTGGTTATGTGGAGCAGGCAGGTGCCGCAGACCTTGATCAATGATGATGGACTGGCGAAGCTAAGCCCGCAGAGCGAGCGCGCCTCGCTGGCGCTGTACGTGTGTACTGCGGAGGAGGCGGCTCGGCACGTGCGGGCGGGGTCGCTGGGCGCGCACGTGCGCCGCGTGCGGGACCTGGCCGGCGCCGCGTTGACTCTCGTCGTCTTCGGCGTCAACGATTACTTCAAGTCGTGCGGCCGCAAGACAATGAACAGCAGCCGGAAACTGATCGGGGAACTTGATTTGGAGCTCGCTATTACCGATTTGCTGGTGACGACCGACTGTGACACGGTCCTGGTGAACTCCTCGAGCGAACTGGCACTGCTGATCGTCCAGCACACCAAGGCGATAGCCGAGGCGCCGTACAAGTAA
Protein
MNKIYKPGECMKYMYVEIHPVLAEAWYMADVSREVTSASAHLITTSSICDPALVMWSRQVPQTLINDDGLAKLSPQSERASLALYVCTAEEAARHVRAGSLGAHVRRVRDLAGAALTLVVFGVNDYFKSCGRKTMNSSRKLIGELDLELAITDLLVTTDCDTVLVNSSSELALLIVQHTKAIAEAPYK
Summary
Uniprot
A0A2A4K6X0
A0A194Q040
A0A194RSB4
A0A3S2PEI2
S4PD80
A0A3S2L718
+ More
A0A2H1VKN1 A0A2H1WLI6 A0A212FM70 E2BNH5 E2AP72 A0A2J7RPL6 A0A0M9AA21 A0A336M418 A0A1B6EFP8 A0A195FGW1 A0A2J7RPK5 A0A336MTC3 A0A2J7RPL3 A0A2A3EP88 F4X397 A0A0J7L200 E9IHL3 A0A088AF59 A0A2Z5TZG6 A0A151WEQ3 A0A026WIC1 A0A151IKY6 A0A310S6K4 A0A139WMN6 A0A139WN59 A0A0L7QYK8 A0A067RC06 A0A151JRI3 A0A1B6CUR5 A0A158NHB2 A0A182MV76 A0A195BEN9 A0A0P6JSN9 A0A2J7RPL4 A0A0A9Y4C6 A0A154PT81 B0WA29 A0A084VSF6 A0A1Y1LWC7 A0A1Q3F2K0 A0A182JEW6 A0A182G1L4 A0A1S4FNY9 A0A182PEL9 A0A1Y1LUS3 A0A182NG93 A0A1Y1LQT7 A0A1Y1LNX2 A0A182W2M3 A0A182R757 A0A1B0C8C0 A0A182K799 A0A182QIX6 A0A3Q0IPC8 A0A093Q4C7 A0A093J4F0 A0A093NY37 A0A2S2QAY4 A0A093JYB2 A0A2I0TRM9 A0A182SGE6 A0A1B6GWG4 A0A182XLZ4 A0A1J1I4G6 W4XD62 A0A087R2W9 A0A182XVE6 A0A0P4W0Q1 A0A091P033 A0A182HTH3 A0A182VG44 A0A182TQD0 A0A091J5L0
A0A2H1VKN1 A0A2H1WLI6 A0A212FM70 E2BNH5 E2AP72 A0A2J7RPL6 A0A0M9AA21 A0A336M418 A0A1B6EFP8 A0A195FGW1 A0A2J7RPK5 A0A336MTC3 A0A2J7RPL3 A0A2A3EP88 F4X397 A0A0J7L200 E9IHL3 A0A088AF59 A0A2Z5TZG6 A0A151WEQ3 A0A026WIC1 A0A151IKY6 A0A310S6K4 A0A139WMN6 A0A139WN59 A0A0L7QYK8 A0A067RC06 A0A151JRI3 A0A1B6CUR5 A0A158NHB2 A0A182MV76 A0A195BEN9 A0A0P6JSN9 A0A2J7RPL4 A0A0A9Y4C6 A0A154PT81 B0WA29 A0A084VSF6 A0A1Y1LWC7 A0A1Q3F2K0 A0A182JEW6 A0A182G1L4 A0A1S4FNY9 A0A182PEL9 A0A1Y1LUS3 A0A182NG93 A0A1Y1LQT7 A0A1Y1LNX2 A0A182W2M3 A0A182R757 A0A1B0C8C0 A0A182K799 A0A182QIX6 A0A3Q0IPC8 A0A093Q4C7 A0A093J4F0 A0A093NY37 A0A2S2QAY4 A0A093JYB2 A0A2I0TRM9 A0A182SGE6 A0A1B6GWG4 A0A182XLZ4 A0A1J1I4G6 W4XD62 A0A087R2W9 A0A182XVE6 A0A0P4W0Q1 A0A091P033 A0A182HTH3 A0A182VG44 A0A182TQD0 A0A091J5L0
Pubmed
EMBL
NWSH01000086
PCG79766.1
KQ459582
KPI98947.1
KQ459700
KPJ20367.1
+ More
RSAL01000071 RVE49095.1 GAIX01003771 JAA88789.1 RSAL01002355 RVE40505.1 ODYU01003090 SOQ41409.1 ODYU01009396 SOQ53792.1 AGBW02007654 OWR54831.1 GL449414 EFN82754.1 GL441481 EFN64745.1 NEVH01001358 PNF42774.1 KQ435706 KOX80287.1 UFQT01000526 SSX24995.1 GEDC01000587 JAS36711.1 KQ981606 KYN39611.1 PNF42772.1 UFQT01001720 SSX31517.1 PNF42770.1 KZ288199 PBC33623.1 GL888609 EGI59078.1 LBMM01001218 KMQ96706.1 GL763289 EFZ19920.1 FX985865 BBA93752.1 KQ983238 KYQ46282.1 KK107207 QOIP01000002 EZA55411.1 RLU26055.1 KQ977151 KYN05258.1 KQ767877 OAD53267.1 KQ971312 KYB29298.1 KYB29297.1 KQ414688 KOC63694.1 KK852605 KDR20403.1 KQ978592 KYN29952.1 GEDC01020072 JAS17226.1 ADTU01000256 AXCM01000235 KQ976509 KYM82652.1 GDUN01000221 JAN95698.1 PNF42773.1 GBHO01017130 GBHO01017128 JAG26474.1 JAG26476.1 KQ435180 KZC14987.1 DS231868 EDS40762.1 ATLV01015947 KE525048 KFB40900.1 GEZM01050880 JAV75327.1 GFDL01013262 JAV21783.1 JXUM01038779 KQ561178 KXJ79461.1 GEZM01050882 GEZM01050878 JAV75326.1 GEZM01050873 JAV75331.1 GEZM01050877 JAV75329.1 AJWK01000333 AXCN02001628 KL671685 KFW83491.1 KK568515 KFW05879.1 KL225043 KFW66905.1 GGMS01005487 MBY74690.1 KL206555 KFV83389.1 KZ507618 PKU36466.1 GECZ01003074 JAS66695.1 CVRI01000037 CRK93273.1 AAGJ04139140 KL226066 KFM07823.1 GDRN01081682 JAI61969.1 KL377626 KFP85050.1 APCN01000528 KK501478 KFP15113.1
RSAL01000071 RVE49095.1 GAIX01003771 JAA88789.1 RSAL01002355 RVE40505.1 ODYU01003090 SOQ41409.1 ODYU01009396 SOQ53792.1 AGBW02007654 OWR54831.1 GL449414 EFN82754.1 GL441481 EFN64745.1 NEVH01001358 PNF42774.1 KQ435706 KOX80287.1 UFQT01000526 SSX24995.1 GEDC01000587 JAS36711.1 KQ981606 KYN39611.1 PNF42772.1 UFQT01001720 SSX31517.1 PNF42770.1 KZ288199 PBC33623.1 GL888609 EGI59078.1 LBMM01001218 KMQ96706.1 GL763289 EFZ19920.1 FX985865 BBA93752.1 KQ983238 KYQ46282.1 KK107207 QOIP01000002 EZA55411.1 RLU26055.1 KQ977151 KYN05258.1 KQ767877 OAD53267.1 KQ971312 KYB29298.1 KYB29297.1 KQ414688 KOC63694.1 KK852605 KDR20403.1 KQ978592 KYN29952.1 GEDC01020072 JAS17226.1 ADTU01000256 AXCM01000235 KQ976509 KYM82652.1 GDUN01000221 JAN95698.1 PNF42773.1 GBHO01017130 GBHO01017128 JAG26474.1 JAG26476.1 KQ435180 KZC14987.1 DS231868 EDS40762.1 ATLV01015947 KE525048 KFB40900.1 GEZM01050880 JAV75327.1 GFDL01013262 JAV21783.1 JXUM01038779 KQ561178 KXJ79461.1 GEZM01050882 GEZM01050878 JAV75326.1 GEZM01050873 JAV75331.1 GEZM01050877 JAV75329.1 AJWK01000333 AXCN02001628 KL671685 KFW83491.1 KK568515 KFW05879.1 KL225043 KFW66905.1 GGMS01005487 MBY74690.1 KL206555 KFV83389.1 KZ507618 PKU36466.1 GECZ01003074 JAS66695.1 CVRI01000037 CRK93273.1 AAGJ04139140 KL226066 KFM07823.1 GDRN01081682 JAI61969.1 KL377626 KFP85050.1 APCN01000528 KK501478 KFP15113.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000283053
UP000007151
UP000008237
+ More
UP000000311 UP000235965 UP000053105 UP000078541 UP000242457 UP000007755 UP000036403 UP000005203 UP000075809 UP000053097 UP000279307 UP000078542 UP000007266 UP000053825 UP000027135 UP000078492 UP000005205 UP000075883 UP000078540 UP000076502 UP000002320 UP000030765 UP000075880 UP000069940 UP000249989 UP000075885 UP000075884 UP000075920 UP000075900 UP000092461 UP000075881 UP000075886 UP000079169 UP000053258 UP000054081 UP000053584 UP000075901 UP000076407 UP000183832 UP000007110 UP000053286 UP000076408 UP000075840 UP000075903 UP000075902 UP000053119
UP000000311 UP000235965 UP000053105 UP000078541 UP000242457 UP000007755 UP000036403 UP000005203 UP000075809 UP000053097 UP000279307 UP000078542 UP000007266 UP000053825 UP000027135 UP000078492 UP000005205 UP000075883 UP000078540 UP000076502 UP000002320 UP000030765 UP000075880 UP000069940 UP000249989 UP000075885 UP000075884 UP000075920 UP000075900 UP000092461 UP000075881 UP000075886 UP000079169 UP000053258 UP000054081 UP000053584 UP000075901 UP000076407 UP000183832 UP000007110 UP000053286 UP000076408 UP000075840 UP000075903 UP000075902 UP000053119
Pfam
PF02732 ERCC4
ProteinModelPortal
A0A2A4K6X0
A0A194Q040
A0A194RSB4
A0A3S2PEI2
S4PD80
A0A3S2L718
+ More
A0A2H1VKN1 A0A2H1WLI6 A0A212FM70 E2BNH5 E2AP72 A0A2J7RPL6 A0A0M9AA21 A0A336M418 A0A1B6EFP8 A0A195FGW1 A0A2J7RPK5 A0A336MTC3 A0A2J7RPL3 A0A2A3EP88 F4X397 A0A0J7L200 E9IHL3 A0A088AF59 A0A2Z5TZG6 A0A151WEQ3 A0A026WIC1 A0A151IKY6 A0A310S6K4 A0A139WMN6 A0A139WN59 A0A0L7QYK8 A0A067RC06 A0A151JRI3 A0A1B6CUR5 A0A158NHB2 A0A182MV76 A0A195BEN9 A0A0P6JSN9 A0A2J7RPL4 A0A0A9Y4C6 A0A154PT81 B0WA29 A0A084VSF6 A0A1Y1LWC7 A0A1Q3F2K0 A0A182JEW6 A0A182G1L4 A0A1S4FNY9 A0A182PEL9 A0A1Y1LUS3 A0A182NG93 A0A1Y1LQT7 A0A1Y1LNX2 A0A182W2M3 A0A182R757 A0A1B0C8C0 A0A182K799 A0A182QIX6 A0A3Q0IPC8 A0A093Q4C7 A0A093J4F0 A0A093NY37 A0A2S2QAY4 A0A093JYB2 A0A2I0TRM9 A0A182SGE6 A0A1B6GWG4 A0A182XLZ4 A0A1J1I4G6 W4XD62 A0A087R2W9 A0A182XVE6 A0A0P4W0Q1 A0A091P033 A0A182HTH3 A0A182VG44 A0A182TQD0 A0A091J5L0
A0A2H1VKN1 A0A2H1WLI6 A0A212FM70 E2BNH5 E2AP72 A0A2J7RPL6 A0A0M9AA21 A0A336M418 A0A1B6EFP8 A0A195FGW1 A0A2J7RPK5 A0A336MTC3 A0A2J7RPL3 A0A2A3EP88 F4X397 A0A0J7L200 E9IHL3 A0A088AF59 A0A2Z5TZG6 A0A151WEQ3 A0A026WIC1 A0A151IKY6 A0A310S6K4 A0A139WMN6 A0A139WN59 A0A0L7QYK8 A0A067RC06 A0A151JRI3 A0A1B6CUR5 A0A158NHB2 A0A182MV76 A0A195BEN9 A0A0P6JSN9 A0A2J7RPL4 A0A0A9Y4C6 A0A154PT81 B0WA29 A0A084VSF6 A0A1Y1LWC7 A0A1Q3F2K0 A0A182JEW6 A0A182G1L4 A0A1S4FNY9 A0A182PEL9 A0A1Y1LUS3 A0A182NG93 A0A1Y1LQT7 A0A1Y1LNX2 A0A182W2M3 A0A182R757 A0A1B0C8C0 A0A182K799 A0A182QIX6 A0A3Q0IPC8 A0A093Q4C7 A0A093J4F0 A0A093NY37 A0A2S2QAY4 A0A093JYB2 A0A2I0TRM9 A0A182SGE6 A0A1B6GWG4 A0A182XLZ4 A0A1J1I4G6 W4XD62 A0A087R2W9 A0A182XVE6 A0A0P4W0Q1 A0A091P033 A0A182HTH3 A0A182VG44 A0A182TQD0 A0A091J5L0
Ontologies
PATHWAY
GO
PANTHER
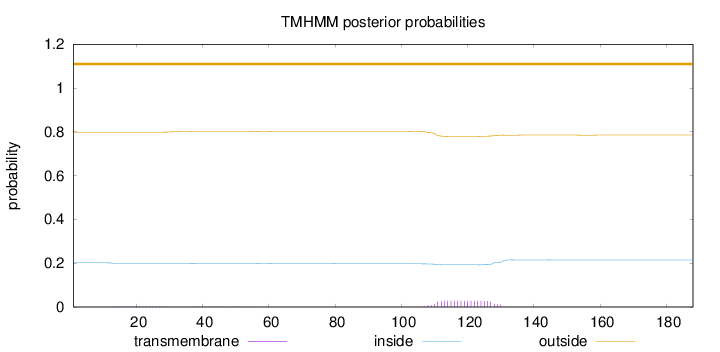
Topology
Length:
188
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.629439999999999
Exp number, first 60 AAs:
0.08415
Total prob of N-in:
0.20167
outside
1 - 188
Population Genetic Test Statistics
Pi
178.791596
Theta
147.126401
Tajima's D
0.643111
CLR
0.06259
CSRT
0.55607219639018
Interpretation
Uncertain
