Pre Gene Modal
BGIBMGA013087
Annotation
PREDICTED:_crossover_junction_endonuclease_EME1-like?_partial_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 3.535
Sequence
CDS
ATGTCGAAGCGCGCTTACGACGAGCAATCCGAATTGTACTTGCGGGGAGAAAACAGGAAATGCGTCACCGTCGACAAACAAGGCAACGGAGTCAGTCGGCTGTGGCAACAGATGATAGCTGTACTGCCACATTCCAGTTTAGAAACGTCGAGAGCATTGTGCGCTAAATATCCAACTCCGCTTGACCTGTATGAGTCCCTTAACTCACCGGATAGTGTTAATGAATTGGCGAACATCGGTGTATCAAGAACCGCTGTCCCGGGTTCAAAGGCGCGTCGCATTGGACCTGAATTTGCCAGGAAATTACACACATTGTTCACAGTCACCGACGGAGATATTTTGTTAGATTGA
Protein
MSKRAYDEQSELYLRGENRKCVTVDKQGNGVSRLWQQMIAVLPHSSLETSRALCAKYPTPLDLYESLNSPDSVNELANIGVSRTAVPGSKARRIGPEFARKLHTLFTVTDGDILLD
Summary
Uniprot
A0A194Q040
S4PD80
A0A194RSB4
A0A2A4K6X0
A0A1E1WKR5
A0A2H1WLI6
+ More
A0A3S2PEI2 A0A212FM70 A0A151IKY6 F4X397 A0A151WEQ3 A0A151JRI3 A0A195BEN9 A0A158NHB2 A0A026WIC1 A0A195FGW1 A0A0M9AA21 E2AP72 E9IHL3 A0A088AF59 A0A0T6B848 E2BNH5 A0A2A3EP88 A0A1B6M529 A0A1Y1LWC7 A0A232F6Q9 A0A1B6CUR5 A0A1Y1LQT7 A0A1Y1LNX2 A0A1Y1LUS3 K7JJF8 A0A1B6L704 A0A154PT81 A0A1B6EFP8 A0A0L7QYK8 A0A1B6IMC5 A0A2H8TQM0 A0A2R7VSU1 J9MAM4 A0A1W4VRD5 J3JUY5 A0A067RC06 A0A023EXB3 A0A336M418 N6TVA2 U4U3N2 A0A336MTC3 A0A0M5J4F6 A0A2J7RPL4 A0A2J7RPK5 B4P833 A0A2J7RPL3 A1Z8C0 Q7JZM5 B3NN50 B4QAN1 B3MIK5 B4KQ86 B4HN03 T1I6B6 A0A2S2QAY4 A0A2Z5TZG6 Q28XK0 B4GG96 A0A0P5T871 B4LKP5 A0A0P4ZMK3 A0A2P8Z785 A0A0P5AHF2 A0A1B0C8C0 A0A0P6F6H5 A0A0P6C877 A0A0P5JXV9 A0A0P4YZZ2 A0A0P4W0Q1 V5FVW2 A0A3B0J033 B4J7U0 A0A1A9ZX59 A0A1B0GEY3 T1GQN5 A0A1A9VY06 A0A3Q0IPC8 A0A423SVG9 A0A1B0BHK4 A0A1I8MVL7 R7UL09 A0A084VSF6 A0A1J1I4G6 A0A444SQW0 A0A444SHJ0 A0A0P5BXY6 A0A182NG93 M7BX91 A0A0L0CDY1 A0A182QIX6
A0A3S2PEI2 A0A212FM70 A0A151IKY6 F4X397 A0A151WEQ3 A0A151JRI3 A0A195BEN9 A0A158NHB2 A0A026WIC1 A0A195FGW1 A0A0M9AA21 E2AP72 E9IHL3 A0A088AF59 A0A0T6B848 E2BNH5 A0A2A3EP88 A0A1B6M529 A0A1Y1LWC7 A0A232F6Q9 A0A1B6CUR5 A0A1Y1LQT7 A0A1Y1LNX2 A0A1Y1LUS3 K7JJF8 A0A1B6L704 A0A154PT81 A0A1B6EFP8 A0A0L7QYK8 A0A1B6IMC5 A0A2H8TQM0 A0A2R7VSU1 J9MAM4 A0A1W4VRD5 J3JUY5 A0A067RC06 A0A023EXB3 A0A336M418 N6TVA2 U4U3N2 A0A336MTC3 A0A0M5J4F6 A0A2J7RPL4 A0A2J7RPK5 B4P833 A0A2J7RPL3 A1Z8C0 Q7JZM5 B3NN50 B4QAN1 B3MIK5 B4KQ86 B4HN03 T1I6B6 A0A2S2QAY4 A0A2Z5TZG6 Q28XK0 B4GG96 A0A0P5T871 B4LKP5 A0A0P4ZMK3 A0A2P8Z785 A0A0P5AHF2 A0A1B0C8C0 A0A0P6F6H5 A0A0P6C877 A0A0P5JXV9 A0A0P4YZZ2 A0A0P4W0Q1 V5FVW2 A0A3B0J033 B4J7U0 A0A1A9ZX59 A0A1B0GEY3 T1GQN5 A0A1A9VY06 A0A3Q0IPC8 A0A423SVG9 A0A1B0BHK4 A0A1I8MVL7 R7UL09 A0A084VSF6 A0A1J1I4G6 A0A444SQW0 A0A444SHJ0 A0A0P5BXY6 A0A182NG93 M7BX91 A0A0L0CDY1 A0A182QIX6
Pubmed
26354079
23622113
22118469
21719571
21347285
24508170
+ More
30249741 20798317 21282665 28004739 28648823 20075255 22516182 24845553 25474469 23537049 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 26760975 15632085 29403074 25315136 23254933 24438588 23624526 26108605
30249741 20798317 21282665 28004739 28648823 20075255 22516182 24845553 25474469 23537049 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 26760975 15632085 29403074 25315136 23254933 24438588 23624526 26108605
EMBL
KQ459582
KPI98947.1
GAIX01003771
JAA88789.1
KQ459700
KPJ20367.1
+ More
NWSH01000086 PCG79766.1 GDQN01003489 JAT87565.1 ODYU01009396 SOQ53792.1 RSAL01000071 RVE49095.1 AGBW02007654 OWR54831.1 KQ977151 KYN05258.1 GL888609 EGI59078.1 KQ983238 KYQ46282.1 KQ978592 KYN29952.1 KQ976509 KYM82652.1 ADTU01000256 KK107207 QOIP01000002 EZA55411.1 RLU26055.1 KQ981606 KYN39611.1 KQ435706 KOX80287.1 GL441481 EFN64745.1 GL763289 EFZ19920.1 LJIG01009210 KRT83536.1 GL449414 EFN82754.1 KZ288199 PBC33623.1 GEBQ01008950 JAT31027.1 GEZM01050880 JAV75327.1 NNAY01000833 OXU26272.1 GEDC01020072 JAS17226.1 GEZM01050873 JAV75331.1 GEZM01050877 JAV75329.1 GEZM01050882 GEZM01050878 JAV75326.1 GEBQ01020521 JAT19456.1 KQ435180 KZC14987.1 GEDC01000587 JAS36711.1 KQ414688 KOC63694.1 GECU01019633 JAS88073.1 GFXV01004475 MBW16280.1 KK854061 PTY10516.1 ABLF02040504 ABLF02040508 BT127050 AEE62012.1 KK852605 KDR20403.1 GBBI01004829 JAC13883.1 UFQT01000526 SSX24995.1 APGK01017247 KB739998 ENN81983.1 KB631984 ERL87672.1 UFQT01001720 SSX31517.1 CP012524 ALC40631.1 NEVH01001358 PNF42773.1 PNF42772.1 CM000158 EDW90080.1 PNF42770.1 AE013599 AAF58752.1 AY071164 AAL48786.2 CH954179 EDV56570.1 CM000362 CM002911 EDX06536.1 KMY92854.1 CH902619 EDV38081.1 CH933808 EDW09214.1 CH480816 EDW47307.1 ACPB03001801 GGMS01005487 MBY74690.1 FX985865 BBA93752.1 CM000071 EAL26316.1 CH479183 EDW35516.1 GDIQ01095586 JAL56140.1 CH940648 EDW61768.1 GDIP01212587 JAJ10815.1 PYGN01000164 PSN52361.1 GDIP01204091 LRGB01001361 JAJ19311.1 KZS12457.1 AJWK01000333 GDIQ01064817 JAN29920.1 GDIP01007346 JAM96369.1 GDIQ01193610 JAK58115.1 GDIP01221135 JAJ02267.1 GDRN01081682 JAI61969.1 GALX01006676 JAB61790.1 OUUW01000001 SPP74154.1 CH916367 EDW01146.1 CCAG010012634 CAQQ02136146 CAQQ02136147 QCYY01002708 ROT68190.1 JXJN01014402 AMQN01001147 KB300094 ELU07229.1 ATLV01015947 KE525048 KFB40900.1 CVRI01000037 CRK93273.1 SAUD01026684 RXG55761.1 SAUD01036743 RXG52793.1 GDIP01182376 JAJ41026.1 KB472301 EMP42471.1 JRES01000637 KNC29704.1 AXCN02001628
NWSH01000086 PCG79766.1 GDQN01003489 JAT87565.1 ODYU01009396 SOQ53792.1 RSAL01000071 RVE49095.1 AGBW02007654 OWR54831.1 KQ977151 KYN05258.1 GL888609 EGI59078.1 KQ983238 KYQ46282.1 KQ978592 KYN29952.1 KQ976509 KYM82652.1 ADTU01000256 KK107207 QOIP01000002 EZA55411.1 RLU26055.1 KQ981606 KYN39611.1 KQ435706 KOX80287.1 GL441481 EFN64745.1 GL763289 EFZ19920.1 LJIG01009210 KRT83536.1 GL449414 EFN82754.1 KZ288199 PBC33623.1 GEBQ01008950 JAT31027.1 GEZM01050880 JAV75327.1 NNAY01000833 OXU26272.1 GEDC01020072 JAS17226.1 GEZM01050873 JAV75331.1 GEZM01050877 JAV75329.1 GEZM01050882 GEZM01050878 JAV75326.1 GEBQ01020521 JAT19456.1 KQ435180 KZC14987.1 GEDC01000587 JAS36711.1 KQ414688 KOC63694.1 GECU01019633 JAS88073.1 GFXV01004475 MBW16280.1 KK854061 PTY10516.1 ABLF02040504 ABLF02040508 BT127050 AEE62012.1 KK852605 KDR20403.1 GBBI01004829 JAC13883.1 UFQT01000526 SSX24995.1 APGK01017247 KB739998 ENN81983.1 KB631984 ERL87672.1 UFQT01001720 SSX31517.1 CP012524 ALC40631.1 NEVH01001358 PNF42773.1 PNF42772.1 CM000158 EDW90080.1 PNF42770.1 AE013599 AAF58752.1 AY071164 AAL48786.2 CH954179 EDV56570.1 CM000362 CM002911 EDX06536.1 KMY92854.1 CH902619 EDV38081.1 CH933808 EDW09214.1 CH480816 EDW47307.1 ACPB03001801 GGMS01005487 MBY74690.1 FX985865 BBA93752.1 CM000071 EAL26316.1 CH479183 EDW35516.1 GDIQ01095586 JAL56140.1 CH940648 EDW61768.1 GDIP01212587 JAJ10815.1 PYGN01000164 PSN52361.1 GDIP01204091 LRGB01001361 JAJ19311.1 KZS12457.1 AJWK01000333 GDIQ01064817 JAN29920.1 GDIP01007346 JAM96369.1 GDIQ01193610 JAK58115.1 GDIP01221135 JAJ02267.1 GDRN01081682 JAI61969.1 GALX01006676 JAB61790.1 OUUW01000001 SPP74154.1 CH916367 EDW01146.1 CCAG010012634 CAQQ02136146 CAQQ02136147 QCYY01002708 ROT68190.1 JXJN01014402 AMQN01001147 KB300094 ELU07229.1 ATLV01015947 KE525048 KFB40900.1 CVRI01000037 CRK93273.1 SAUD01026684 RXG55761.1 SAUD01036743 RXG52793.1 GDIP01182376 JAJ41026.1 KB472301 EMP42471.1 JRES01000637 KNC29704.1 AXCN02001628
Proteomes
UP000053268
UP000053240
UP000218220
UP000283053
UP000007151
UP000078542
+ More
UP000007755 UP000075809 UP000078492 UP000078540 UP000005205 UP000053097 UP000279307 UP000078541 UP000053105 UP000000311 UP000005203 UP000008237 UP000242457 UP000215335 UP000002358 UP000076502 UP000053825 UP000007819 UP000192221 UP000027135 UP000019118 UP000030742 UP000092553 UP000235965 UP000002282 UP000000803 UP000008711 UP000000304 UP000007801 UP000009192 UP000001292 UP000015103 UP000001819 UP000008744 UP000008792 UP000245037 UP000076858 UP000092461 UP000268350 UP000001070 UP000092445 UP000092444 UP000015102 UP000078200 UP000079169 UP000283509 UP000092460 UP000095301 UP000014760 UP000030765 UP000183832 UP000288706 UP000075884 UP000031443 UP000037069 UP000075886
UP000007755 UP000075809 UP000078492 UP000078540 UP000005205 UP000053097 UP000279307 UP000078541 UP000053105 UP000000311 UP000005203 UP000008237 UP000242457 UP000215335 UP000002358 UP000076502 UP000053825 UP000007819 UP000192221 UP000027135 UP000019118 UP000030742 UP000092553 UP000235965 UP000002282 UP000000803 UP000008711 UP000000304 UP000007801 UP000009192 UP000001292 UP000015103 UP000001819 UP000008744 UP000008792 UP000245037 UP000076858 UP000092461 UP000268350 UP000001070 UP000092445 UP000092444 UP000015102 UP000078200 UP000079169 UP000283509 UP000092460 UP000095301 UP000014760 UP000030765 UP000183832 UP000288706 UP000075884 UP000031443 UP000037069 UP000075886
Pfam
PF02732 ERCC4
ProteinModelPortal
A0A194Q040
S4PD80
A0A194RSB4
A0A2A4K6X0
A0A1E1WKR5
A0A2H1WLI6
+ More
A0A3S2PEI2 A0A212FM70 A0A151IKY6 F4X397 A0A151WEQ3 A0A151JRI3 A0A195BEN9 A0A158NHB2 A0A026WIC1 A0A195FGW1 A0A0M9AA21 E2AP72 E9IHL3 A0A088AF59 A0A0T6B848 E2BNH5 A0A2A3EP88 A0A1B6M529 A0A1Y1LWC7 A0A232F6Q9 A0A1B6CUR5 A0A1Y1LQT7 A0A1Y1LNX2 A0A1Y1LUS3 K7JJF8 A0A1B6L704 A0A154PT81 A0A1B6EFP8 A0A0L7QYK8 A0A1B6IMC5 A0A2H8TQM0 A0A2R7VSU1 J9MAM4 A0A1W4VRD5 J3JUY5 A0A067RC06 A0A023EXB3 A0A336M418 N6TVA2 U4U3N2 A0A336MTC3 A0A0M5J4F6 A0A2J7RPL4 A0A2J7RPK5 B4P833 A0A2J7RPL3 A1Z8C0 Q7JZM5 B3NN50 B4QAN1 B3MIK5 B4KQ86 B4HN03 T1I6B6 A0A2S2QAY4 A0A2Z5TZG6 Q28XK0 B4GG96 A0A0P5T871 B4LKP5 A0A0P4ZMK3 A0A2P8Z785 A0A0P5AHF2 A0A1B0C8C0 A0A0P6F6H5 A0A0P6C877 A0A0P5JXV9 A0A0P4YZZ2 A0A0P4W0Q1 V5FVW2 A0A3B0J033 B4J7U0 A0A1A9ZX59 A0A1B0GEY3 T1GQN5 A0A1A9VY06 A0A3Q0IPC8 A0A423SVG9 A0A1B0BHK4 A0A1I8MVL7 R7UL09 A0A084VSF6 A0A1J1I4G6 A0A444SQW0 A0A444SHJ0 A0A0P5BXY6 A0A182NG93 M7BX91 A0A0L0CDY1 A0A182QIX6
A0A3S2PEI2 A0A212FM70 A0A151IKY6 F4X397 A0A151WEQ3 A0A151JRI3 A0A195BEN9 A0A158NHB2 A0A026WIC1 A0A195FGW1 A0A0M9AA21 E2AP72 E9IHL3 A0A088AF59 A0A0T6B848 E2BNH5 A0A2A3EP88 A0A1B6M529 A0A1Y1LWC7 A0A232F6Q9 A0A1B6CUR5 A0A1Y1LQT7 A0A1Y1LNX2 A0A1Y1LUS3 K7JJF8 A0A1B6L704 A0A154PT81 A0A1B6EFP8 A0A0L7QYK8 A0A1B6IMC5 A0A2H8TQM0 A0A2R7VSU1 J9MAM4 A0A1W4VRD5 J3JUY5 A0A067RC06 A0A023EXB3 A0A336M418 N6TVA2 U4U3N2 A0A336MTC3 A0A0M5J4F6 A0A2J7RPL4 A0A2J7RPK5 B4P833 A0A2J7RPL3 A1Z8C0 Q7JZM5 B3NN50 B4QAN1 B3MIK5 B4KQ86 B4HN03 T1I6B6 A0A2S2QAY4 A0A2Z5TZG6 Q28XK0 B4GG96 A0A0P5T871 B4LKP5 A0A0P4ZMK3 A0A2P8Z785 A0A0P5AHF2 A0A1B0C8C0 A0A0P6F6H5 A0A0P6C877 A0A0P5JXV9 A0A0P4YZZ2 A0A0P4W0Q1 V5FVW2 A0A3B0J033 B4J7U0 A0A1A9ZX59 A0A1B0GEY3 T1GQN5 A0A1A9VY06 A0A3Q0IPC8 A0A423SVG9 A0A1B0BHK4 A0A1I8MVL7 R7UL09 A0A084VSF6 A0A1J1I4G6 A0A444SQW0 A0A444SHJ0 A0A0P5BXY6 A0A182NG93 M7BX91 A0A0L0CDY1 A0A182QIX6
PDB
2ZIX
E-value=1.06097e-07,
Score=127
Ontologies
PANTHER
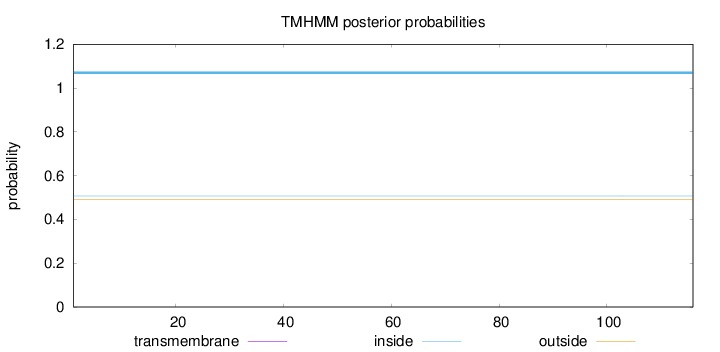
Topology
Length:
116
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000940000000000001
Exp number, first 60 AAs:
0.00058
Total prob of N-in:
0.50737
inside
1 - 116
Population Genetic Test Statistics
Pi
163.89645
Theta
134.739248
Tajima's D
0.268565
CLR
11.929371
CSRT
0.448627568621569
Interpretation
Uncertain
