Gene
KWMTBOMO09403
Pre Gene Modal
BGIBMGA001193
Annotation
PREDICTED:_uncharacterized_protein_LOC106135401_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 1.719
Sequence
CDS
ATGCATAAAGACAGCCAGATACAGCCGTCGATACTGCCTCGGGTGCCGTGTGCTGTAACTGCTGATGCTCATTCACAAACCCAGAGTGTTATCCATACTCTGGACGATTTGGCATCTTACAAAAATAAAATTCCTGTGCTTAAGCACATCCCTAAGGGGGCCAGGAATTTAGTCGCAGGAAAGCTCAGAGTGATTATGGACGGCTGCATAAACAATAACGGTGTGGCGGACTGGGTTTCCTTGTTGTCATTTTCTTACACGACTCTGAGAATTCCAGATCGAGGTGACCCCCGTTCGCTTACTGCAAAGGTTAAGGAGAATGCAAACAATAATAGCCCGTATTTTTTAGAGGACTTCAAACACAAGCCACGATCCATTGTCAAAAGAATCGAAGCCAAGGTTCATGAGGGTGATTTGCGTGGAGCTGTCCGTCTTTTAGTGTCGGAGGATTCTTTAGCACCTTTTAACGATGAGACCTTAAGCGCATTACGTGATAAACATCCTACTCCTTTTCGTCCCTTATCATTGCCGCCAGCGCCTGATTCTAGTTGCCCTTTTTTGACTGTCAACTCGGAGGATGTTGCAGACGCAATTAGTTCCTTTTATAGCGGCTCTGCACCTGGCCTTGATGGTCTACGTCCAAACCATTTAAAGGAGCTCATTTCCCCTTCCGCCGGTGAAAATGGTGTTCGTCTATTGCACTCAATTACAGACTTATGCAATTTCATACTAAGAGGTACGGTTAATGTACAGGTGTGTCCTTATTTGTACGGAGGTAGCTTATGCGCTCTAACTAAGAAGGACGGCGGTGTGAGACCAATTGCGGTGGGGTCTGTGCTGCGTCGTCTCGCTGCCAAGCTTGGGTGTCGGTCTGTAAGGAATGAAATGGCCTCGTACTTGCAGCCCCACCAGCTGGGCTTCGGGACTGCTCTTGGGTGTGAGGCCGCCATTCATGCTACCCGTGCATTTGCTATGAGCAGGGAGAGCTGTAACAGTGCAATTGTAAAGCTTGATATAAGGAATGCTTTTAATAGCTTAGAGAGAGATATAATTTTAAATGAAATCAAGGAAAAGATTCCGGCTTTATACCCATTCTTGTATCAGTGTTACAGCTCTTCCAGCAAGTTATTCTTTATGAAATCTTCCATTGACTCAGAGGTAGGTGCCCAGCAGGGCGATCCTCTTGGCCCATTAATTTTTAGTCTTGCTATCCACAGCATTATAAAGACCGTCAAGTCACCTTTAAACCTATGGTATTTAGATGACGGCACAATAGGTGGTCCCCCAGATTCCGTGCTAGACGACATCAGACTTTTATTTCCGAAGTTGCGAGATGTCGGCTTGGAGCAAGCGTAG
Protein
MHKDSQIQPSILPRVPCAVTADAHSQTQSVIHTLDDLASYKNKIPVLKHIPKGARNLVAGKLRVIMDGCINNNGVADWVSLLSFSYTTLRIPDRGDPRSLTAKVKENANNNSPYFLEDFKHKPRSIVKRIEAKVHEGDLRGAVRLLVSEDSLAPFNDETLSALRDKHPTPFRPLSLPPAPDSSCPFLTVNSEDVADAISSFYSGSAPGLDGLRPNHLKELISPSAGENGVRLLHSITDLCNFILRGTVNVQVCPYLYGGSLCALTKKDGGVRPIAVGSVLRRLAAKLGCRSVRNEMASYLQPHQLGFGTALGCEAAIHATRAFAMSRESCNSAIVKLDIRNAFNSLERDIILNEIKEKIPALYPFLYQCYSSSSKLFFMKSSIDSEVGAQQGDPLGPLIFSLAIHSIIKTVKSPLNLWYLDDGTIGGPPDSVLDDIRLLFPKLRDVGLEQA
Summary
Similarity
Belongs to the amiloride-sensitive sodium channel (TC 1.A.6) family.
Uniprot
A0A1S3DQS3
A0A2P8ZLV7
A0A194R3A6
A0A1W0X6D1
A0A226DSQ4
A0A226ENY2
+ More
A0A1D1V0I0 A0A1D1VL73 A0A226DV40 A0A226F2H2 A0A1D1V0B4 A0A2V3IC56 A0A226DD92 A0A1Y1MJ73 A0A1D1V341 A0A1X7SSY7 A0A2B7ZX51 A0A226DXH2 A0A226EVZ6 A0A226D463 A0A1D1W821 A0A1X7TNR1 A0A226CYT8 A0A1Q9DZ07 A0A1Q9E7W5 A0A2K3NHH2 A0A251UV27 A0A1Q9C9N2 A0A1Q9DL84 A0A251RPE8 A0A251U8Z7 A0A1Q9CLQ1 A0A251SLN7 X6NKH2 X6LZW2 X6LZS6 A0A1X7UXU0 X6LXW7
A0A1D1V0I0 A0A1D1VL73 A0A226DV40 A0A226F2H2 A0A1D1V0B4 A0A2V3IC56 A0A226DD92 A0A1Y1MJ73 A0A1D1V341 A0A1X7SSY7 A0A2B7ZX51 A0A226DXH2 A0A226EVZ6 A0A226D463 A0A1D1W821 A0A1X7TNR1 A0A226CYT8 A0A1Q9DZ07 A0A1Q9E7W5 A0A2K3NHH2 A0A251UV27 A0A1Q9C9N2 A0A1Q9DL84 A0A251RPE8 A0A251U8Z7 A0A1Q9CLQ1 A0A251SLN7 X6NKH2 X6LZW2 X6LZS6 A0A1X7UXU0 X6LXW7
EMBL
PYGN01000020
PSN57485.1
KQ460779
KPJ12288.1
MTYJ01000015
OQV22928.1
+ More
LNIX01000012 OXA48253.1 LNIX01000003 OXA58286.1 BDGG01000002 GAU92213.1 BDGG01000007 GAV01566.1 LNIX01000011 OXA48561.1 LNIX01000001 OXA63637.1 BDGG01000003 GAU95299.1 NBIV01000529 PXF39673.1 LNIX01000023 OXA43090.1 GEZM01032301 JAV84680.1 GAU95300.1 PDHN01000780 PGH37981.1 LNIX01000010 OXA49381.1 OXA61805.1 LNIX01000038 OXA39608.1 BDGG01000023 GAV09476.1 LNIX01000053 OXA37708.1 LSRX01000328 OLQ00411.1 LSRX01000234 OLQ03518.1 ASHM01021424 PNY02492.1 CM007893 OTG26899.1 LSRX01001462 OLP79640.1 LSRX01000486 OLP95909.1 CM007906 OTF86222.1 CM007897 OTG19252.1 LSRX01001088 OLP83851.1 CM007903 OTF99774.1 ASPP01007617 ETO26815.1 ASPP01026421 ETO07174.1 ASPP01027314 ETO06245.1 ASPP01027610 ETO05987.1
LNIX01000012 OXA48253.1 LNIX01000003 OXA58286.1 BDGG01000002 GAU92213.1 BDGG01000007 GAV01566.1 LNIX01000011 OXA48561.1 LNIX01000001 OXA63637.1 BDGG01000003 GAU95299.1 NBIV01000529 PXF39673.1 LNIX01000023 OXA43090.1 GEZM01032301 JAV84680.1 GAU95300.1 PDHN01000780 PGH37981.1 LNIX01000010 OXA49381.1 OXA61805.1 LNIX01000038 OXA39608.1 BDGG01000023 GAV09476.1 LNIX01000053 OXA37708.1 LSRX01000328 OLQ00411.1 LSRX01000234 OLQ03518.1 ASHM01021424 PNY02492.1 CM007893 OTG26899.1 LSRX01001462 OLP79640.1 LSRX01000486 OLP95909.1 CM007906 OTF86222.1 CM007897 OTG19252.1 LSRX01001088 OLP83851.1 CM007903 OTF99774.1 ASPP01007617 ETO26815.1 ASPP01026421 ETO07174.1 ASPP01027314 ETO06245.1 ASPP01027610 ETO05987.1
Proteomes
Pfam
Interpro
IPR000477
RT_dom
+ More
IPR013087 Znf_C2H2_type
IPR036388 WH-like_DNA-bd_sf
IPR011011 Znf_FYVE_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR019787 Znf_PHD-finger
IPR019786 Zinc_finger_PHD-type_CS
IPR003150 DNA-bd_RFX
IPR036236 Znf_C2H2_sf
IPR001873 ENaC
IPR004875 DDE_SF_endonuclease_dom
IPR020903 ENaC_CS
IPR036390 WH_DNA-bd_sf
IPR017452 GPCR_Rhodpsn_7TM
IPR000276 GPCR_Rhodpsn
IPR039779 RFX-like
IPR036322 WD40_repeat_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR037151 AlkB-like_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR004263 Exostosin
IPR013087 Znf_C2H2_type
IPR036388 WH-like_DNA-bd_sf
IPR011011 Znf_FYVE_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR019787 Znf_PHD-finger
IPR019786 Zinc_finger_PHD-type_CS
IPR003150 DNA-bd_RFX
IPR036236 Znf_C2H2_sf
IPR001873 ENaC
IPR004875 DDE_SF_endonuclease_dom
IPR020903 ENaC_CS
IPR036390 WH_DNA-bd_sf
IPR017452 GPCR_Rhodpsn_7TM
IPR000276 GPCR_Rhodpsn
IPR039779 RFX-like
IPR036322 WD40_repeat_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR037151 AlkB-like_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR004263 Exostosin
Gene 3D
ProteinModelPortal
A0A1S3DQS3
A0A2P8ZLV7
A0A194R3A6
A0A1W0X6D1
A0A226DSQ4
A0A226ENY2
+ More
A0A1D1V0I0 A0A1D1VL73 A0A226DV40 A0A226F2H2 A0A1D1V0B4 A0A2V3IC56 A0A226DD92 A0A1Y1MJ73 A0A1D1V341 A0A1X7SSY7 A0A2B7ZX51 A0A226DXH2 A0A226EVZ6 A0A226D463 A0A1D1W821 A0A1X7TNR1 A0A226CYT8 A0A1Q9DZ07 A0A1Q9E7W5 A0A2K3NHH2 A0A251UV27 A0A1Q9C9N2 A0A1Q9DL84 A0A251RPE8 A0A251U8Z7 A0A1Q9CLQ1 A0A251SLN7 X6NKH2 X6LZW2 X6LZS6 A0A1X7UXU0 X6LXW7
A0A1D1V0I0 A0A1D1VL73 A0A226DV40 A0A226F2H2 A0A1D1V0B4 A0A2V3IC56 A0A226DD92 A0A1Y1MJ73 A0A1D1V341 A0A1X7SSY7 A0A2B7ZX51 A0A226DXH2 A0A226EVZ6 A0A226D463 A0A1D1W821 A0A1X7TNR1 A0A226CYT8 A0A1Q9DZ07 A0A1Q9E7W5 A0A2K3NHH2 A0A251UV27 A0A1Q9C9N2 A0A1Q9DL84 A0A251RPE8 A0A251U8Z7 A0A1Q9CLQ1 A0A251SLN7 X6NKH2 X6LZW2 X6LZS6 A0A1X7UXU0 X6LXW7
Ontologies
KEGG
GO
Topology
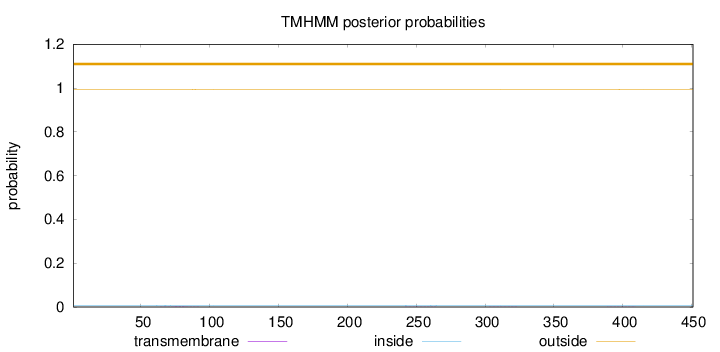
Length:
451
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04791
Exp number, first 60 AAs:
7e-05
Total prob of N-in:
0.00768
outside
1 - 451
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
0
CSRT
0
Interpretation
Uncertain
