Gene
KWMTBOMO09387 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013098
Annotation
PREDICTED:_uncharacterized_protein_LOC106107572_isoform_X1_[Papilio_polytes]
Full name
Poxin
Location in the cell
Cytoplasmic Reliability : 1.696 Mitochondrial Reliability : 1.195
Sequence
CDS
ATGTCTAAGCGCACAGTACTAAACGAGAATTACAAGGGCATAGTGGAGTCGATGAGTATTCCTGCTGAAATACACGAGAGAAACGGCAAGAAGTACGCGAGCGTGGGGTCCATACTGCCGATACACTGCTGCCCGCCCGAAGAGCTGGAGCGACGCGCTGAGAGCACGCACCACTACTGCGGCGTGTTCACCGACGAACTGCTGGCGCCGCTCGAGGAACTGGCCTACGTTAGACTCGACGAGAATACTGCTGAGAAGGTCTTCATAAACAGAGCCAAGCGCATACTGATAGTGTCAAGCGACGGGCACCTGGCGCAGTGGCGGTGCGCGCCGACCTTCGAGTCTGCGAACCGGTACATCGCCGGCACGCCCATCGTCGACCAGCGCGGGGGCGTCATCTCCGTCGTTGTGGCGAAGAAAAACAACCACTATGCCGTCTCCTCCTTCGAGGGTGAAGGCGGATACTTTGAGAGCACACAAAACTGGAAGGTGGTTGAACCGGCTGCAGGCGGCTATGCGTACGGGGAGTTGACTTTCCCGAGCCGCACCGCCCTCCGCGAGCACGTGGCGGGGCTGCGCGGGGGCGCGGGAGCGTGGGGGGACGCCGTGCCCGTGCTGCGGGGGGGCACGTCGCCGCGCCTCGCGCTGGTGCTGGACGGCAGGCAGCTCGCACACTACTACCTGCACAACGTTATTGTGGACGTAGAATACTTGTAA
Protein
MSKRTVLNENYKGIVESMSIPAEIHERNGKKYASVGSILPIHCCPPEELERRAESTHHYCGVFTDELLAPLEELAYVRLDENTAEKVFINRAKRILIVSSDGHLAQWRCAPTFESANRYIAGTPIVDQRGGVISVVVAKKNNHYAVSSFEGEGGYFESTQNWKVVEPAAGGYAYGELTFPSRTALREHVAGLRGGAGAWGDAVPVLRGGTSPRLALVLDGRQLAHYYLHNVIVDVEYL
Summary
Description
Nuclease that cleaves 2',3'-cGAMP.
Catalytic Activity
2'-3'-cGAMP + H2O = Gp(2'-5')Ap(3') + H(+)
Similarity
Belongs to the poxin family. Highly divergent.
Keywords
Complete proteome
Hydrolase
Nuclease
Reference proteome
Feature
chain Poxin
Uniprot
EC Number
3.1.-.-
EMBL
Proteomes
PRIDE
ProteinModelPortal
Ontologies
GO
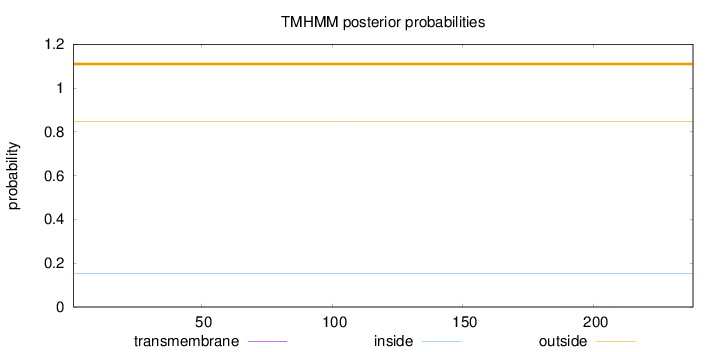
Topology
Length:
238
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00612999999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.15183
outside
1 - 238
Population Genetic Test Statistics
Pi
221.409591
Theta
196.564387
Tajima's D
0.389165
CLR
0.117671
CSRT
0.483525823708815
Interpretation
Uncertain
