Pre Gene Modal
BGIBMGA012759
Annotation
PREDICTED:_uncharacterized_protein_LOC106109791_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 1.068 Mitochondrial Reliability : 1.46
Sequence
CDS
ATGGGAAGTCATCAAAGTCATGATAAAGCGGACGGGAGTGGGAGCGGTACGAGTACACCCCGAGGCTCCCAGCGGGGTGCCGGGTCGCGATCAGGCTCCGGGGGTGACCTGCAGGGGCTGGCCGAGCGACAAGAGAAACCTCCACTTTCTATGGTGCCAGTTGAGAAACTAGGAAAATTACTGGCACAAATATCACAGAAAGAAGATGGAATCAACGGAGTCACAGAGAATTCTTTTAAGAAATACTTATTCCCGATGTACCCGGAGCTGGCACACCACTTCTACAAGTACATCCACAGAATCGGCAAGTGCAAGAACAAACACATACCTCTGTCTACGTTCAGAATGCAGTGCGAGAAGATACTCAGCATGCTAGACGACGCGGTCATCATTGAGACATACGTCAGAATGTTCAGTATGGAAACAGACGAGAATAACATGACACCAGACGGTCTCAGGAACATGCTGTACACCAGCTACAAGTTGTCCATGGACCACTATCCAGAAGGACCACAAACTTGTCTCATGATCAACCAGACGCTATCGGCCGTGGTGGACGGCTGCTTCAACAAGAAGGACGTGCACAGCGTCGGCTTCGTGGTGCGCTGGCTCGAGGAGCACTGCCACCGGATCATATTCCCGGTCCACAGGTACTGCGTGCACCTGCTGGCGACGCGGCACCGCGACATGGAGGCGCACGTGGAGTCGCGCGGCGCGGGCGCGGGGCTGGAGCTGGCCACGCCGGTGCTGGAGCGGGGCGGGGCGGCGCGGGGCTCGGCGCTGCTGCCGGTGTCGGCCGCGTGGCTGCTGGCCGCCGCCGCGCCGCCCCGCTACTCGCGCCCCGCCGCGCCGCCCGCGCCCTCCGCCGGCTGGCTGGTGGCGCGCCTGGTGTGCGCCGTGCCCTCGCACTGGACGCCGCTCTACTCCAGCTCCGACCAGGGCCTCGCCGCCAACAGGTTCCTGCACCACACGCTGGCGTACCGCGGCCCCACCGTGGTGCTGGCGCGCGGCGGGCCGCTGCTGGTGGCGCTGTGCTGCCCCGCGGAGTGGCGCGACTCGCACTGCTACTGGGGGGACGCCGACTGCGCGCTGCTGCAGCTGCTGCCCACGTTCTCGCTGATAGAGCGCGCGCCGAAGATGATATACCTGAACACGTCGATCCGCGGCTACCCCAAGGGTCTGCGCGCCGGCTCCGACCCGCGCCGCCCCGCGCTCGCCATCGCAGAGGGCTTCGACTCCTTCACCTTCAACGGCGCGCCCTACTCACTCGACAGCATTGAGGTGTGGGGCTGCGGGGACCAGGCATCGAGGGAAACCCAACTGGAGATCAAAAAGTGGCAAATACGGGAAGCGGAAAAACAAAGACAAGTGAAGCTGTCGGCGGCGGACTGGATGGAACACCCGGACCGCTACCTGCTGGAGCTGGCGGGCCGCCCGCAGTACAACAACTCCGCCAGCTAG
Protein
MGSHQSHDKADGSGSGTSTPRGSQRGAGSRSGSGGDLQGLAERQEKPPLSMVPVEKLGKLLAQISQKEDGINGVTENSFKKYLFPMYPELAHHFYKYIHRIGKCKNKHIPLSTFRMQCEKILSMLDDAVIIETYVRMFSMETDENNMTPDGLRNMLYTSYKLSMDHYPEGPQTCLMINQTLSAVVDGCFNKKDVHSVGFVVRWLEEHCHRIIFPVHRYCVHLLATRHRDMEAHVESRGAGAGLELATPVLERGGAARGSALLPVSAAWLLAAAAPPRYSRPAAPPAPSAGWLVARLVCAVPSHWTPLYSSSDQGLAANRFLHHTLAYRGPTVVLARGGPLLVALCCPAEWRDSHCYWGDADCALLQLLPTFSLIERAPKMIYLNTSIRGYPKGLRAGSDPRRPALAIAEGFDSFTFNGAPYSLDSIEVWGCGDQASRETQLEIKKWQIREAEKQRQVKLSAADWMEHPDRYLLELAGRPQYNNSAS
Summary
Uniprot
A0A2A4K8L7
A0A2A4K8X6
A0A2A4K9C3
A0A194Q0X8
A0A2W1BP84
A0A212FME0
+ More
A0A2A4K8C4 A0A0L7LFL8 V5GHT7 A0A1Y1NHJ1 D2A5C2 Q0IEJ0 A0A1S4FQB3 K7J2T5 A0A336LLU8 A0A336ME30 B0WMK8 A0A2M3Z0J5 A0A2M3ZFV9 A0A0C9RVN1 A0A232F3F1 A0A2M4BJF3 A0A2M4BJM9 A0A2J7PX51 A0A1B6DBB8 A0A310SK31 A0A067RI68 A0A182FTL6 E0VH45 J9JQ62 A0A3L8DE22 A0A026WUH5 A0A195BAZ8 A0A158NK53 F4W8E9 E2A0A2 A0A2S2QKT6 A0A195FB63 A0A1Q3FFK7 A0A151X2U0 A0A195EJ00 A0A1Q3FFN5 A0A0L7QSI0 E9IDW8 E2BTZ5 A0A1B6IM64 A0A1B6FL81 A0A154PHW1 A0A2A3EEP5 A0A195C2T0 A0A087ZST2 A0A0M9A7R8 A0A1L8EGQ9 A0A1I8NZH4 A0A0K8UHX9 B3N8L5 B4NYQ4 W8BSZ9 A0A034WP86 B3MPC2 A0A1J1HH36 D3TMR2 B4HWD9 B4Q8A9 Q9VL57 A0A2P8ZJK6 A0A0A1XI39 A0A1W4UFL4 A0A2M4A8M2 A0A2M4A8H5 A0A1B0ABA7 A0A1I8MQI1 A0A240SWZ0 U4U7M6 A0A2J7PX56 A0A1L8EB39 A0A2H8U0L1 A0A1A9VE67 A0A1I8NZF1 A0A0K8WI03 X2J7M7 A0A0A1WTQ9 A0A1B0C1D0 A0A1A9YNY6
A0A2A4K8C4 A0A0L7LFL8 V5GHT7 A0A1Y1NHJ1 D2A5C2 Q0IEJ0 A0A1S4FQB3 K7J2T5 A0A336LLU8 A0A336ME30 B0WMK8 A0A2M3Z0J5 A0A2M3ZFV9 A0A0C9RVN1 A0A232F3F1 A0A2M4BJF3 A0A2M4BJM9 A0A2J7PX51 A0A1B6DBB8 A0A310SK31 A0A067RI68 A0A182FTL6 E0VH45 J9JQ62 A0A3L8DE22 A0A026WUH5 A0A195BAZ8 A0A158NK53 F4W8E9 E2A0A2 A0A2S2QKT6 A0A195FB63 A0A1Q3FFK7 A0A151X2U0 A0A195EJ00 A0A1Q3FFN5 A0A0L7QSI0 E9IDW8 E2BTZ5 A0A1B6IM64 A0A1B6FL81 A0A154PHW1 A0A2A3EEP5 A0A195C2T0 A0A087ZST2 A0A0M9A7R8 A0A1L8EGQ9 A0A1I8NZH4 A0A0K8UHX9 B3N8L5 B4NYQ4 W8BSZ9 A0A034WP86 B3MPC2 A0A1J1HH36 D3TMR2 B4HWD9 B4Q8A9 Q9VL57 A0A2P8ZJK6 A0A0A1XI39 A0A1W4UFL4 A0A2M4A8M2 A0A2M4A8H5 A0A1B0ABA7 A0A1I8MQI1 A0A240SWZ0 U4U7M6 A0A2J7PX56 A0A1L8EB39 A0A2H8U0L1 A0A1A9VE67 A0A1I8NZF1 A0A0K8WI03 X2J7M7 A0A0A1WTQ9 A0A1B0C1D0 A0A1A9YNY6
Pubmed
26354079
28756777
22118469
26227816
28004739
18362917
+ More
19820115 17510324 20075255 28648823 24845553 20566863 30249741 24508170 21347285 21719571 20798317 21282665 17994087 18057021 17550304 24495485 25348373 20353571 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 29403074 25830018 25315136 23537049
19820115 17510324 20075255 28648823 24845553 20566863 30249741 24508170 21347285 21719571 20798317 21282665 17994087 18057021 17550304 24495485 25348373 20353571 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 29403074 25830018 25315136 23537049
EMBL
NWSH01000045
PCG80254.1
PCG80253.1
PCG80252.1
KQ459582
KPI98968.1
+ More
KZ149997 PZC75395.1 AGBW02007654 OWR54849.1 PCG80256.1 JTDY01001334 KOB74164.1 GALX01004867 JAB63599.1 GEZM01002468 JAV97239.1 KQ971345 EFA05347.1 CH477655 EAT37720.1 UFQS01000041 UFQT01000041 SSW98309.1 SSX18695.1 UFQS01000766 UFQT01000766 SSX06717.1 SSX27063.1 DS231998 EDS31079.1 GGFM01001265 MBW22016.1 GGFM01006683 MBW27434.1 GBYB01013020 JAG82787.1 NNAY01001153 OXU24973.1 GGFJ01004021 MBW53162.1 GGFJ01004020 MBW53161.1 NEVH01020862 PNF20911.1 GEDC01014301 JAS22997.1 KQ760505 OAD60072.1 KK852619 KDR20105.1 DS235158 EEB12701.1 ABLF02025277 QOIP01000009 RLU18403.1 KK107119 EZA58764.1 KQ976537 KYM81395.1 ADTU01018700 GL887908 EGI69440.1 GL435563 EFN73137.1 GGMS01009131 MBY78334.1 KQ981693 KYN37621.1 GFDL01008712 JAV26333.1 KQ982579 KYQ54572.1 KQ978881 KYN27839.1 GFDL01008685 JAV26360.1 KQ414756 KOC61597.1 GL762535 EFZ21269.1 GL450531 EFN80894.1 GECU01019693 JAS88013.1 GECZ01019006 JAS50763.1 KQ434912 KZC11402.1 KZ288266 PBC30207.1 KQ978344 KYM94900.1 KQ435718 KOX78957.1 GFDG01000955 JAV17844.1 GDHF01026050 JAI26264.1 CH954177 EDV58438.1 KQS70403.1 KQS70404.1 CM000157 EDW88718.1 KRJ97987.1 KRJ97988.1 GAMC01004258 JAC02298.1 GAKP01002947 JAC56005.1 CH902620 EDV32241.1 KPU73856.1 CVRI01000004 CRK87336.1 EZ422714 ADD18990.1 CH480818 EDW52334.1 CM000361 CM002910 EDX04431.1 KMY89375.1 AE014134 BT004863 AAF52839.1 AAO45219.1 PYGN01000035 PSN56687.1 GBXI01003707 JAD10585.1 GGFK01003794 MBW37115.1 GGFK01003748 MBW37069.1 CCAG010007529 KB631843 ERL86626.1 PNF20909.1 GFDG01003026 JAV15773.1 GFXV01007133 MBW18938.1 GDHF01001809 JAI50505.1 AHN54304.1 GBXI01012256 JAD02036.1 JXJN01023987
KZ149997 PZC75395.1 AGBW02007654 OWR54849.1 PCG80256.1 JTDY01001334 KOB74164.1 GALX01004867 JAB63599.1 GEZM01002468 JAV97239.1 KQ971345 EFA05347.1 CH477655 EAT37720.1 UFQS01000041 UFQT01000041 SSW98309.1 SSX18695.1 UFQS01000766 UFQT01000766 SSX06717.1 SSX27063.1 DS231998 EDS31079.1 GGFM01001265 MBW22016.1 GGFM01006683 MBW27434.1 GBYB01013020 JAG82787.1 NNAY01001153 OXU24973.1 GGFJ01004021 MBW53162.1 GGFJ01004020 MBW53161.1 NEVH01020862 PNF20911.1 GEDC01014301 JAS22997.1 KQ760505 OAD60072.1 KK852619 KDR20105.1 DS235158 EEB12701.1 ABLF02025277 QOIP01000009 RLU18403.1 KK107119 EZA58764.1 KQ976537 KYM81395.1 ADTU01018700 GL887908 EGI69440.1 GL435563 EFN73137.1 GGMS01009131 MBY78334.1 KQ981693 KYN37621.1 GFDL01008712 JAV26333.1 KQ982579 KYQ54572.1 KQ978881 KYN27839.1 GFDL01008685 JAV26360.1 KQ414756 KOC61597.1 GL762535 EFZ21269.1 GL450531 EFN80894.1 GECU01019693 JAS88013.1 GECZ01019006 JAS50763.1 KQ434912 KZC11402.1 KZ288266 PBC30207.1 KQ978344 KYM94900.1 KQ435718 KOX78957.1 GFDG01000955 JAV17844.1 GDHF01026050 JAI26264.1 CH954177 EDV58438.1 KQS70403.1 KQS70404.1 CM000157 EDW88718.1 KRJ97987.1 KRJ97988.1 GAMC01004258 JAC02298.1 GAKP01002947 JAC56005.1 CH902620 EDV32241.1 KPU73856.1 CVRI01000004 CRK87336.1 EZ422714 ADD18990.1 CH480818 EDW52334.1 CM000361 CM002910 EDX04431.1 KMY89375.1 AE014134 BT004863 AAF52839.1 AAO45219.1 PYGN01000035 PSN56687.1 GBXI01003707 JAD10585.1 GGFK01003794 MBW37115.1 GGFK01003748 MBW37069.1 CCAG010007529 KB631843 ERL86626.1 PNF20909.1 GFDG01003026 JAV15773.1 GFXV01007133 MBW18938.1 GDHF01001809 JAI50505.1 AHN54304.1 GBXI01012256 JAD02036.1 JXJN01023987
Proteomes
UP000218220
UP000053268
UP000007151
UP000037510
UP000007266
UP000008820
+ More
UP000002358 UP000002320 UP000215335 UP000235965 UP000027135 UP000069272 UP000009046 UP000007819 UP000279307 UP000053097 UP000078540 UP000005205 UP000007755 UP000000311 UP000078541 UP000075809 UP000078492 UP000053825 UP000008237 UP000076502 UP000242457 UP000078542 UP000005203 UP000053105 UP000095300 UP000008711 UP000002282 UP000007801 UP000183832 UP000001292 UP000000304 UP000000803 UP000245037 UP000192221 UP000092445 UP000095301 UP000092444 UP000030742 UP000078200 UP000092460 UP000092443
UP000002358 UP000002320 UP000215335 UP000235965 UP000027135 UP000069272 UP000009046 UP000007819 UP000279307 UP000053097 UP000078540 UP000005205 UP000007755 UP000000311 UP000078541 UP000075809 UP000078492 UP000053825 UP000008237 UP000076502 UP000242457 UP000078542 UP000005203 UP000053105 UP000095300 UP000008711 UP000002282 UP000007801 UP000183832 UP000001292 UP000000304 UP000000803 UP000245037 UP000192221 UP000092445 UP000095301 UP000092444 UP000030742 UP000078200 UP000092460 UP000092443
Interpro
Gene 3D
ProteinModelPortal
A0A2A4K8L7
A0A2A4K8X6
A0A2A4K9C3
A0A194Q0X8
A0A2W1BP84
A0A212FME0
+ More
A0A2A4K8C4 A0A0L7LFL8 V5GHT7 A0A1Y1NHJ1 D2A5C2 Q0IEJ0 A0A1S4FQB3 K7J2T5 A0A336LLU8 A0A336ME30 B0WMK8 A0A2M3Z0J5 A0A2M3ZFV9 A0A0C9RVN1 A0A232F3F1 A0A2M4BJF3 A0A2M4BJM9 A0A2J7PX51 A0A1B6DBB8 A0A310SK31 A0A067RI68 A0A182FTL6 E0VH45 J9JQ62 A0A3L8DE22 A0A026WUH5 A0A195BAZ8 A0A158NK53 F4W8E9 E2A0A2 A0A2S2QKT6 A0A195FB63 A0A1Q3FFK7 A0A151X2U0 A0A195EJ00 A0A1Q3FFN5 A0A0L7QSI0 E9IDW8 E2BTZ5 A0A1B6IM64 A0A1B6FL81 A0A154PHW1 A0A2A3EEP5 A0A195C2T0 A0A087ZST2 A0A0M9A7R8 A0A1L8EGQ9 A0A1I8NZH4 A0A0K8UHX9 B3N8L5 B4NYQ4 W8BSZ9 A0A034WP86 B3MPC2 A0A1J1HH36 D3TMR2 B4HWD9 B4Q8A9 Q9VL57 A0A2P8ZJK6 A0A0A1XI39 A0A1W4UFL4 A0A2M4A8M2 A0A2M4A8H5 A0A1B0ABA7 A0A1I8MQI1 A0A240SWZ0 U4U7M6 A0A2J7PX56 A0A1L8EB39 A0A2H8U0L1 A0A1A9VE67 A0A1I8NZF1 A0A0K8WI03 X2J7M7 A0A0A1WTQ9 A0A1B0C1D0 A0A1A9YNY6
A0A2A4K8C4 A0A0L7LFL8 V5GHT7 A0A1Y1NHJ1 D2A5C2 Q0IEJ0 A0A1S4FQB3 K7J2T5 A0A336LLU8 A0A336ME30 B0WMK8 A0A2M3Z0J5 A0A2M3ZFV9 A0A0C9RVN1 A0A232F3F1 A0A2M4BJF3 A0A2M4BJM9 A0A2J7PX51 A0A1B6DBB8 A0A310SK31 A0A067RI68 A0A182FTL6 E0VH45 J9JQ62 A0A3L8DE22 A0A026WUH5 A0A195BAZ8 A0A158NK53 F4W8E9 E2A0A2 A0A2S2QKT6 A0A195FB63 A0A1Q3FFK7 A0A151X2U0 A0A195EJ00 A0A1Q3FFN5 A0A0L7QSI0 E9IDW8 E2BTZ5 A0A1B6IM64 A0A1B6FL81 A0A154PHW1 A0A2A3EEP5 A0A195C2T0 A0A087ZST2 A0A0M9A7R8 A0A1L8EGQ9 A0A1I8NZH4 A0A0K8UHX9 B3N8L5 B4NYQ4 W8BSZ9 A0A034WP86 B3MPC2 A0A1J1HH36 D3TMR2 B4HWD9 B4Q8A9 Q9VL57 A0A2P8ZJK6 A0A0A1XI39 A0A1W4UFL4 A0A2M4A8M2 A0A2M4A8H5 A0A1B0ABA7 A0A1I8MQI1 A0A240SWZ0 U4U7M6 A0A2J7PX56 A0A1L8EB39 A0A2H8U0L1 A0A1A9VE67 A0A1I8NZF1 A0A0K8WI03 X2J7M7 A0A0A1WTQ9 A0A1B0C1D0 A0A1A9YNY6
Ontologies
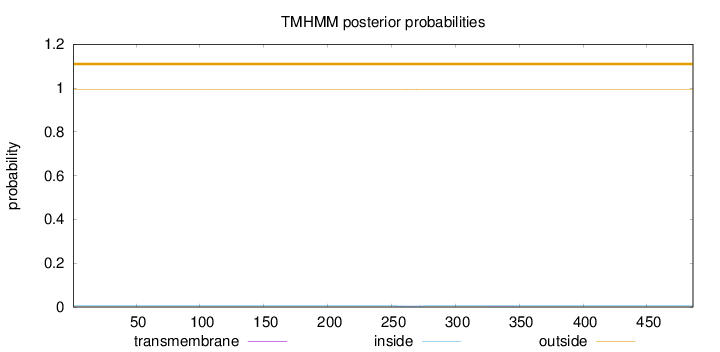
Topology
Length:
486
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02857
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00558
outside
1 - 486
Population Genetic Test Statistics
Pi
284.08937
Theta
198.381024
Tajima's D
1.522756
CLR
0.16592
CSRT
0.788460576971151
Interpretation
Uncertain
