Gene
KWMTBOMO09374
Pre Gene Modal
BGIBMGA013107
Annotation
PREDICTED:_uncharacterized_protein_LOC101744804_isoform_X1_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.828
Sequence
CDS
ATGCGCGGGTGGTGGAGCGGCTGGACGAGGTCGCGGGTGCGGTGGCTGGCGGCGCTGGTGTGCTGGGGCGGGCTGGCGGCGCTCGCCCTGCCGCGCCTGCAGCCGCCGCCGCCCGCGCACCGCGCCCGCCCCGCGCCCCCCTCGCGCCTCCTGCTGCAGTCCGAGCCCGCCGCCTCCACGCCGCCCGCCCCCCTCCACGCCCTGCACGCGCTCCCAGCCACCGACACTCGCGTGCTCACCGAGGAGGAGCTGCGGGCCGACGCCGAGCGGCGCCTGCCCTCGCTGCCTCTGGCGTACTGGCACAGGCACAAGGACGACAAGAACAAGTACTACAAGAAGAAGGAGTGCGCGCCCTTCCCCTCCATCTACGACCTCGAGTTCCACAACACCTACTGGCAGACGATGCACTCGAGCGAGGGCGCGTTCCACCTGTACGGCGCGTACATGGACCGGCGCAACACGTCGCGCATCGGGCCCGCCGTGCGCGTGCTCGCCGTGCACGACCGCATCAAGCCCACCATCGCCACGCACTGCCAGCTCTGGTTCGAGGAGCGCGACCAGCCCGTGGTCGTGCGCAACCTCGAGTACAAGTACGTGTGGAACAGCAAGTGGGGTAACTACCGCGACCGCGTGCTGCAGCCGTACCTGCTGGCCTGCGTGCTGCCGGCCGAGGTGCGGGACCTGGTGCCCGCCTCCGTGTCGCTCGTGGAGAACCCCTGCGACCGCGCGACTAACAACCTGCGCGTCCACTACGACGCGCCGCCGACGGGGGGGCGCAAGGAGTTCGCGGTCTGCGTCAAGGGCCTAGACTTCCTGCACGAGGACCTCTCGGTGCGCCTCGTCGAGTGGATCGAGCTCGTGCGCTTGCTCGGGGCCGACAAGATCTTTTTCTATGAGCTTCAGGTCCATCCGAATATCACAAAAGTTCTCGACTACTATGCGAACCTCGGCGCAGTCACGGTGACGCCGATCACTCTGCCCGGAGGCCAACCGAACCTGCCGGGTCTCCAGCACATGTACCTCAAGAAGAAGACGACCCACAAGCGACAGATGGAGTTGATACCGTACAACGATTGCTTGTATCGGCACATGTACGAGTACCGGTGGCTGGCGCTCCTGGACATCGACGAGGTGATCGTGCCGCTGGAGGACGGCGGCTGGAGCGGGCTCATGCGGCGCGCCCTGCCGCTGTCGGCGCCGGCACCGGGCAAGCCGCCGCGCTCCTCGTACCACGCCTCCAACCTGTACTTCCTGGACTCGCTGCGGCACGAGCACGGCTGGGAGGAGGGCGTGCCGCGCTACATGCACATGCTGCAGCACGTGTACCGCACGCGCAACTTCACCAAGCCTGGCCAGTACGTGAAGGCGTTCCACGAGACGGACCGCGTGCTGGCGCTGCACAACCACTTCCCGCTGGCGTGCCTGGGCGGCGCGTGCTCCTCCTACGCGCTGGAGACCAGGCACGCGCGGCTGCAGCACTACCGCGCGGACTGCGTCACGGCGCTCAGCAAGACCTGCGCCGAGCTGAGAGCGCAGCCCGAGCGGGACACCTCGCTGTGGCGTTGGAAAGACGTGCTGGTGGCGCGTGCGGACGAGGCGCTCACCAAGCTTGGCTTCCTGCCGCCACACCGGTGA
Protein
MRGWWSGWTRSRVRWLAALVCWGGLAALALPRLQPPPPAHRARPAPPSRLLLQSEPAASTPPAPLHALHALPATDTRVLTEEELRADAERRLPSLPLAYWHRHKDDKNKYYKKKECAPFPSIYDLEFHNTYWQTMHSSEGAFHLYGAYMDRRNTSRIGPAVRVLAVHDRIKPTIATHCQLWFEERDQPVVVRNLEYKYVWNSKWGNYRDRVLQPYLLACVLPAEVRDLVPASVSLVENPCDRATNNLRVHYDAPPTGGRKEFAVCVKGLDFLHEDLSVRLVEWIELVRLLGADKIFFYELQVHPNITKVLDYYANLGAVTVTPITLPGGQPNLPGLQHMYLKKKTTHKRQMELIPYNDCLYRHMYEYRWLALLDIDEVIVPLEDGGWSGLMRRALPLSAPAPGKPPRSSYHASNLYFLDSLRHEHGWEEGVPRYMHMLQHVYRTRNFTKPGQYVKAFHETDRVLALHNHFPLACLGGACSSYALETRHARLQHYRADCVTALSKTCAELRAQPERDTSLWRWKDVLVARADEALTKLGFLPPHR
Summary
Uniprot
H9JU95
A0A0N0PCP8
A0A194Q1V7
A0A2H1WBD4
A0A0L7LDQ4
A0A2A4JI57
+ More
A0A1B6DZ50 U4USI8 N6SVQ6 A0A2J7PMH3 A0A2S2PRH8 A0A2P8Y4E0 A0A1S3CXK5 D2A5K9 A0A1Y1JXE1 A0A067RD35 J9JY70 A0A0L7RCA7 A0A1B6FCX5 A0A026WCB8 A0A2S2RAA6 A0A1L8E1L6 E2C7C8 A0A154P7X9 A0A1L8E1F5 A0A1L8E1K1 A0A1B6MRJ5 T1HIZ0 A0A0M8ZVX9 A0A1B0CGA0 A0A1W4WSS6 A0A0A9YC52 A0A2A3ESN5 V9ILB0 A0A087ZNN9 A0A195FUN5 A0A195BD98 A0A158P3F4 A0A2P8YGB6 F4WM62 A0A1B0D8J1 A0A310SF26 A0A336M9K8 A0A195DCD8 A0A151IGF0 A0A023F4D9 A0A2R7VUM0 A0A151WFI4 E9IXY4 E2AQ10 A0A182GGC8 Q17B39 B0WWJ1 A0A0C9RWP3 A0A034VHN2 A0A0K8VWW0 A0A0A1WID7 A0A2M4CQN4 W5JEA9 W8BDK7 A0A084VAI8 A0A182JJH6 A0A182QQC0 A0A182VRI5 T1PCY6 E0VMI1 A0A182X2Q6 A0A182VBQ6 A0A182TKF3 A0A182F6V1 T1PL98 A0A182NFJ7 A0A0N7ZD56 A0A0P4WCC5 A0A182XV93 A0A182HGS5 A0A182KMB2 Q7PY98 A0A182K777 B3MYF9 Q9V3U1 A0A423T3E5 M9PGK1 B4I9A0 A0A182MJ23 A0A182P9T1 A0A1W4VE30 B4PXK9 B4GTR2 A0A1I8PWV4 B3P9D9 B4L3U0 A0A182RNZ8 A0A3B0KKK1 Q29IX3 B4JKT8 A0A1A9W206 A0A3B0KR43
A0A1B6DZ50 U4USI8 N6SVQ6 A0A2J7PMH3 A0A2S2PRH8 A0A2P8Y4E0 A0A1S3CXK5 D2A5K9 A0A1Y1JXE1 A0A067RD35 J9JY70 A0A0L7RCA7 A0A1B6FCX5 A0A026WCB8 A0A2S2RAA6 A0A1L8E1L6 E2C7C8 A0A154P7X9 A0A1L8E1F5 A0A1L8E1K1 A0A1B6MRJ5 T1HIZ0 A0A0M8ZVX9 A0A1B0CGA0 A0A1W4WSS6 A0A0A9YC52 A0A2A3ESN5 V9ILB0 A0A087ZNN9 A0A195FUN5 A0A195BD98 A0A158P3F4 A0A2P8YGB6 F4WM62 A0A1B0D8J1 A0A310SF26 A0A336M9K8 A0A195DCD8 A0A151IGF0 A0A023F4D9 A0A2R7VUM0 A0A151WFI4 E9IXY4 E2AQ10 A0A182GGC8 Q17B39 B0WWJ1 A0A0C9RWP3 A0A034VHN2 A0A0K8VWW0 A0A0A1WID7 A0A2M4CQN4 W5JEA9 W8BDK7 A0A084VAI8 A0A182JJH6 A0A182QQC0 A0A182VRI5 T1PCY6 E0VMI1 A0A182X2Q6 A0A182VBQ6 A0A182TKF3 A0A182F6V1 T1PL98 A0A182NFJ7 A0A0N7ZD56 A0A0P4WCC5 A0A182XV93 A0A182HGS5 A0A182KMB2 Q7PY98 A0A182K777 B3MYF9 Q9V3U1 A0A423T3E5 M9PGK1 B4I9A0 A0A182MJ23 A0A182P9T1 A0A1W4VE30 B4PXK9 B4GTR2 A0A1I8PWV4 B3P9D9 B4L3U0 A0A182RNZ8 A0A3B0KKK1 Q29IX3 B4JKT8 A0A1A9W206 A0A3B0KR43
Pubmed
19121390
26354079
26227816
23537049
29403074
18362917
+ More
19820115 28004739 24845553 24508170 30249741 20798317 25401762 26823975 21347285 21719571 25474469 21282665 26483478 17510324 25348373 25830018 20920257 23761445 24495485 24438588 25315136 20566863 25244985 20966253 12364791 14747013 17210077 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085
19820115 28004739 24845553 24508170 30249741 20798317 25401762 26823975 21347285 21719571 25474469 21282665 26483478 17510324 25348373 25830018 20920257 23761445 24495485 24438588 25315136 20566863 25244985 20966253 12364791 14747013 17210077 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085
EMBL
BABH01004289
BABH01004290
BABH01004291
KQ460635
KPJ13430.1
KQ459582
+ More
KPI98979.1 ODYU01007524 SOQ50373.1 JTDY01001558 KOB73535.1 NWSH01001304 PCG71775.1 GEDC01006340 JAS30958.1 KB632429 ERL95483.1 APGK01054650 APGK01054651 KB741251 ENN71839.1 NEVH01023991 PNF17527.1 GGMR01019404 MBY32023.1 PYGN01000949 PSN39044.1 KQ971345 EFA05385.1 GEZM01102952 GEZM01102951 GEZM01102950 GEZM01102949 JAV51686.1 KK852715 KDR17871.1 ABLF02034075 KQ414616 KOC68468.1 GECZ01030926 GECZ01021700 GECZ01011113 JAS38843.1 JAS48069.1 JAS58656.1 KK107293 QOIP01000005 EZA53321.1 RLU22947.1 GGMS01017079 MBY86282.1 GFDF01001622 JAV12462.1 GL453369 EFN76115.1 KQ434839 KZC08025.1 GFDF01001557 JAV12527.1 GFDF01001558 JAV12526.1 GEBQ01001472 JAT38505.1 ACPB03006498 KQ435847 KOX71126.1 AJWK01010890 GBHO01013850 GDHC01020840 JAG29754.1 JAP97788.1 KZ288186 PBC34765.1 JR049744 AEY61059.1 KQ981264 KYN44002.1 KQ976519 KYM82182.1 ADTU01001324 PYGN01000616 PSN43298.1 GL888217 EGI64706.1 AJVK01012652 KQ769179 OAD52905.1 UFQT01000636 SSX25991.1 KQ980989 KYN10575.1 KQ977726 KYN00304.1 GBBI01002322 JAC16390.1 KK854054 PTY10390.1 KQ983219 KYQ46545.1 GL766836 EFZ14565.1 GL441645 EFN64497.1 JXUM01061675 KQ562163 KXJ76536.1 CH477326 EAT43472.1 DS232148 EDS36083.1 GBYB01013463 GBYB01013465 JAG83230.1 JAG83232.1 GAKP01016933 JAC42019.1 GDHF01027925 GDHF01015659 GDHF01008970 JAI24389.1 JAI36655.1 JAI43344.1 GBXI01015861 JAC98430.1 GGFL01003397 MBW67575.1 ADMH02001382 ETN62692.1 GAMC01018697 GAMC01018696 JAB87859.1 ATLV01004124 KE524190 KFB34982.1 AXCN02000209 KA646529 KA649540 KA649541 AFP61158.1 DS235312 EEB14587.1 KA649542 AFP64171.1 GDRN01052334 JAI66312.1 GDRN01052335 JAI66311.1 APCN01005944 APCN01005945 AAAB01008987 EAA01177.4 CH902632 EDV32653.2 AE014298 AY069777 AY599409 AL132651 AAF45582.1 AAL39922.1 AAT09452.1 AGB94965.1 CAB59574.1 QCYY01002364 ROT70984.1 AGB94964.1 CH480825 EDW43781.1 AXCM01009966 CM000162 EDX00862.2 CH479190 EDW25932.1 CH954183 EDV45435.2 CH933810 EDW07218.2 OUUW01000018 SPP89100.1 CH379063 EAL32530.2 CH916370 EDW00191.1 SPP89099.1
KPI98979.1 ODYU01007524 SOQ50373.1 JTDY01001558 KOB73535.1 NWSH01001304 PCG71775.1 GEDC01006340 JAS30958.1 KB632429 ERL95483.1 APGK01054650 APGK01054651 KB741251 ENN71839.1 NEVH01023991 PNF17527.1 GGMR01019404 MBY32023.1 PYGN01000949 PSN39044.1 KQ971345 EFA05385.1 GEZM01102952 GEZM01102951 GEZM01102950 GEZM01102949 JAV51686.1 KK852715 KDR17871.1 ABLF02034075 KQ414616 KOC68468.1 GECZ01030926 GECZ01021700 GECZ01011113 JAS38843.1 JAS48069.1 JAS58656.1 KK107293 QOIP01000005 EZA53321.1 RLU22947.1 GGMS01017079 MBY86282.1 GFDF01001622 JAV12462.1 GL453369 EFN76115.1 KQ434839 KZC08025.1 GFDF01001557 JAV12527.1 GFDF01001558 JAV12526.1 GEBQ01001472 JAT38505.1 ACPB03006498 KQ435847 KOX71126.1 AJWK01010890 GBHO01013850 GDHC01020840 JAG29754.1 JAP97788.1 KZ288186 PBC34765.1 JR049744 AEY61059.1 KQ981264 KYN44002.1 KQ976519 KYM82182.1 ADTU01001324 PYGN01000616 PSN43298.1 GL888217 EGI64706.1 AJVK01012652 KQ769179 OAD52905.1 UFQT01000636 SSX25991.1 KQ980989 KYN10575.1 KQ977726 KYN00304.1 GBBI01002322 JAC16390.1 KK854054 PTY10390.1 KQ983219 KYQ46545.1 GL766836 EFZ14565.1 GL441645 EFN64497.1 JXUM01061675 KQ562163 KXJ76536.1 CH477326 EAT43472.1 DS232148 EDS36083.1 GBYB01013463 GBYB01013465 JAG83230.1 JAG83232.1 GAKP01016933 JAC42019.1 GDHF01027925 GDHF01015659 GDHF01008970 JAI24389.1 JAI36655.1 JAI43344.1 GBXI01015861 JAC98430.1 GGFL01003397 MBW67575.1 ADMH02001382 ETN62692.1 GAMC01018697 GAMC01018696 JAB87859.1 ATLV01004124 KE524190 KFB34982.1 AXCN02000209 KA646529 KA649540 KA649541 AFP61158.1 DS235312 EEB14587.1 KA649542 AFP64171.1 GDRN01052334 JAI66312.1 GDRN01052335 JAI66311.1 APCN01005944 APCN01005945 AAAB01008987 EAA01177.4 CH902632 EDV32653.2 AE014298 AY069777 AY599409 AL132651 AAF45582.1 AAL39922.1 AAT09452.1 AGB94965.1 CAB59574.1 QCYY01002364 ROT70984.1 AGB94964.1 CH480825 EDW43781.1 AXCM01009966 CM000162 EDX00862.2 CH479190 EDW25932.1 CH954183 EDV45435.2 CH933810 EDW07218.2 OUUW01000018 SPP89100.1 CH379063 EAL32530.2 CH916370 EDW00191.1 SPP89099.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000037510
UP000218220
UP000030742
+ More
UP000019118 UP000235965 UP000245037 UP000079169 UP000007266 UP000027135 UP000007819 UP000053825 UP000053097 UP000279307 UP000008237 UP000076502 UP000015103 UP000053105 UP000092461 UP000192223 UP000242457 UP000005203 UP000078541 UP000078540 UP000005205 UP000007755 UP000092462 UP000078492 UP000078542 UP000075809 UP000000311 UP000069940 UP000249989 UP000008820 UP000002320 UP000000673 UP000030765 UP000075880 UP000075886 UP000075920 UP000095301 UP000009046 UP000076407 UP000075903 UP000075902 UP000069272 UP000075884 UP000076408 UP000075840 UP000075882 UP000007062 UP000075881 UP000007801 UP000000803 UP000283509 UP000001292 UP000075883 UP000075885 UP000192221 UP000002282 UP000008744 UP000095300 UP000008711 UP000009192 UP000075900 UP000268350 UP000001819 UP000001070 UP000091820
UP000019118 UP000235965 UP000245037 UP000079169 UP000007266 UP000027135 UP000007819 UP000053825 UP000053097 UP000279307 UP000008237 UP000076502 UP000015103 UP000053105 UP000092461 UP000192223 UP000242457 UP000005203 UP000078541 UP000078540 UP000005205 UP000007755 UP000092462 UP000078492 UP000078542 UP000075809 UP000000311 UP000069940 UP000249989 UP000008820 UP000002320 UP000000673 UP000030765 UP000075880 UP000075886 UP000075920 UP000095301 UP000009046 UP000076407 UP000075903 UP000075902 UP000069272 UP000075884 UP000076408 UP000075840 UP000075882 UP000007062 UP000075881 UP000007801 UP000000803 UP000283509 UP000001292 UP000075883 UP000075885 UP000192221 UP000002282 UP000008744 UP000095300 UP000008711 UP000009192 UP000075900 UP000268350 UP000001819 UP000001070 UP000091820
Pfam
PF01697 Glyco_transf_92
Interpro
IPR008166
Glyco_transf_92
ProteinModelPortal
H9JU95
A0A0N0PCP8
A0A194Q1V7
A0A2H1WBD4
A0A0L7LDQ4
A0A2A4JI57
+ More
A0A1B6DZ50 U4USI8 N6SVQ6 A0A2J7PMH3 A0A2S2PRH8 A0A2P8Y4E0 A0A1S3CXK5 D2A5K9 A0A1Y1JXE1 A0A067RD35 J9JY70 A0A0L7RCA7 A0A1B6FCX5 A0A026WCB8 A0A2S2RAA6 A0A1L8E1L6 E2C7C8 A0A154P7X9 A0A1L8E1F5 A0A1L8E1K1 A0A1B6MRJ5 T1HIZ0 A0A0M8ZVX9 A0A1B0CGA0 A0A1W4WSS6 A0A0A9YC52 A0A2A3ESN5 V9ILB0 A0A087ZNN9 A0A195FUN5 A0A195BD98 A0A158P3F4 A0A2P8YGB6 F4WM62 A0A1B0D8J1 A0A310SF26 A0A336M9K8 A0A195DCD8 A0A151IGF0 A0A023F4D9 A0A2R7VUM0 A0A151WFI4 E9IXY4 E2AQ10 A0A182GGC8 Q17B39 B0WWJ1 A0A0C9RWP3 A0A034VHN2 A0A0K8VWW0 A0A0A1WID7 A0A2M4CQN4 W5JEA9 W8BDK7 A0A084VAI8 A0A182JJH6 A0A182QQC0 A0A182VRI5 T1PCY6 E0VMI1 A0A182X2Q6 A0A182VBQ6 A0A182TKF3 A0A182F6V1 T1PL98 A0A182NFJ7 A0A0N7ZD56 A0A0P4WCC5 A0A182XV93 A0A182HGS5 A0A182KMB2 Q7PY98 A0A182K777 B3MYF9 Q9V3U1 A0A423T3E5 M9PGK1 B4I9A0 A0A182MJ23 A0A182P9T1 A0A1W4VE30 B4PXK9 B4GTR2 A0A1I8PWV4 B3P9D9 B4L3U0 A0A182RNZ8 A0A3B0KKK1 Q29IX3 B4JKT8 A0A1A9W206 A0A3B0KR43
A0A1B6DZ50 U4USI8 N6SVQ6 A0A2J7PMH3 A0A2S2PRH8 A0A2P8Y4E0 A0A1S3CXK5 D2A5K9 A0A1Y1JXE1 A0A067RD35 J9JY70 A0A0L7RCA7 A0A1B6FCX5 A0A026WCB8 A0A2S2RAA6 A0A1L8E1L6 E2C7C8 A0A154P7X9 A0A1L8E1F5 A0A1L8E1K1 A0A1B6MRJ5 T1HIZ0 A0A0M8ZVX9 A0A1B0CGA0 A0A1W4WSS6 A0A0A9YC52 A0A2A3ESN5 V9ILB0 A0A087ZNN9 A0A195FUN5 A0A195BD98 A0A158P3F4 A0A2P8YGB6 F4WM62 A0A1B0D8J1 A0A310SF26 A0A336M9K8 A0A195DCD8 A0A151IGF0 A0A023F4D9 A0A2R7VUM0 A0A151WFI4 E9IXY4 E2AQ10 A0A182GGC8 Q17B39 B0WWJ1 A0A0C9RWP3 A0A034VHN2 A0A0K8VWW0 A0A0A1WID7 A0A2M4CQN4 W5JEA9 W8BDK7 A0A084VAI8 A0A182JJH6 A0A182QQC0 A0A182VRI5 T1PCY6 E0VMI1 A0A182X2Q6 A0A182VBQ6 A0A182TKF3 A0A182F6V1 T1PL98 A0A182NFJ7 A0A0N7ZD56 A0A0P4WCC5 A0A182XV93 A0A182HGS5 A0A182KMB2 Q7PY98 A0A182K777 B3MYF9 Q9V3U1 A0A423T3E5 M9PGK1 B4I9A0 A0A182MJ23 A0A182P9T1 A0A1W4VE30 B4PXK9 B4GTR2 A0A1I8PWV4 B3P9D9 B4L3U0 A0A182RNZ8 A0A3B0KKK1 Q29IX3 B4JKT8 A0A1A9W206 A0A3B0KR43
Ontologies
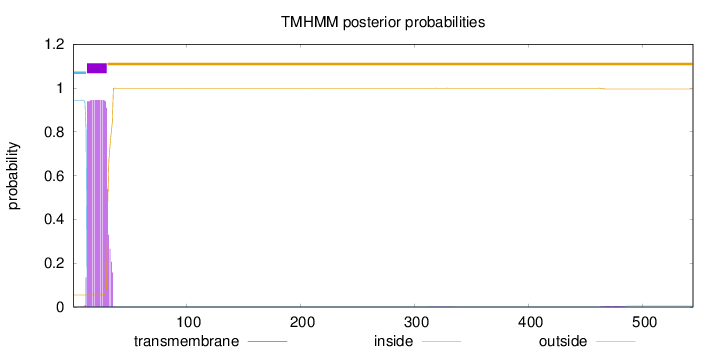
Topology
Length:
544
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
18.66952
Exp number, first 60 AAs:
18.58562
Total prob of N-in:
0.94478
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 30
outside
31 - 544
Population Genetic Test Statistics
Pi
7.184482
Theta
8.892537
Tajima's D
-0.730803
CLR
0
CSRT
0.191490425478726
Interpretation
Uncertain
