Gene
KWMTBOMO09372
Pre Gene Modal
BGIBMGA002197
Annotation
PREDICTED:_uncharacterized_protein_LOC101740270_isoform_X2_[Bombyx_mori]
Full name
Guanylate cyclase
Location in the cell
Mitochondrial Reliability : 1.712 Nuclear Reliability : 1.042
Sequence
CDS
ATGCGCGCGCTGCCCGCCGTGCTCACCACGCTCACCGCGCTCACCGTGCTCACCGTGCTCACCACACTCACCGCGCTCAGCGTGCACGCACACGCCGGCCCGCCTGCCCTGCTAGTGCTGCGTTCCGCCTGCTTTCGACAGGACTTGCCTCGTTTGCATCCAATACCTCGCGCTACGCTTCTCTCTCATCAAGAGACATGCGATGTTGGTTGGAACGAGCGGGTAACTCGGAGGGCCGTGTCGACTCTGACCGACGGCGGGCTGGTGGAGATCATCGGATGGCCCGGGGCCGCGGTATGCGAGTCCATATCCGTAACGGCGAGTGTACTGCGGCGGCCGCAGCCAGCCTCGTGCCTGAGGCTGAAGGCGCAGGCCGCTGCGGTGGCGGAGCTGGCGCGGAGGCTGCGCTGGTCGCGCGCCCTGCTCCCTCGCGCGCCGCCCTCCTCCGACTGCGAGGCCGCTCTGCGCGAGTTAACGCTCTTCCTGCTAGAGACCGGTCTGCTTGTCCGGCGCGCGATGCTCTCCGCCCGCGCCTTCGCCGACCGACCGGAAGTTCTCATTTTATGCGTATCGGTGGCTGCGAAGATCGGCAGCAAGATAGAGTCGATGTTAAAAGATACCGATGTTGTTATTTTGCTTCTACATTTTGAAGAACTCATGCGAGACGCTACTGACTTTGCTAATAAAGACATCGAAAACTTTAAGACAATGAAATCTGAATCAATAAAAGCATCAAAAGTCAATTACTCGTCAGACTCTACTAGCGAACGTGAAATTAATGATCACAAATTGAATCATGAAATGGTACAAAATTTAAATTCACCTCTAAGTTTCTACAACATTCATCGAAAAATGCAAATCAGGTCTGGCAAGTCCTCTATGAATTCAACTGAATTAAGTACAAATCAATTAATTGTTCCGTTCACGAAACCCAGTGAGGGAAATCAAACTGAGCCTGCGAATATTGTTCACGAAGGTACTCGGTCATTAGACGACCGTCGCGATGCTCGCGGAACAACTATGAGCACCAAACACGCAGAAATTGAATCTAGTCAAACGCTGCAAATTTCTAAAAAAGATTTAAGATACCTTGAGATTTTTTCTCATTTTAACGACTTGTTTCCTGGGTGGCTTTTAATTCTAAGTGAAAATATATTCGTCGAGGAACATCTGACGTCTGATTCCATCTCTGAGAAAGTCACAAAAGTGTTGGAGAGCGTTGAAGAGAGCGAGCGCGCGCGCCAGCAGAGCGTGCAGAGGAGGTTCCTCCTGTACGTGCGCAGCGGCGACCGCTCGGCGTGGAGCCGCGCCGCCACGCTCGCCACCAACCTCACGCGCGCGCTCGACGGCGACCGCCCGTGGGCTGCCACCGTCGCACTGCACGACTGGCACGCTCTGGAGCGCTGGAACGGAACCAAGCCCGAAGACGAGCGCGACCATTGGAACGGCTCCAACATCCTGGCGACCGTGCTGGGAGCGGGCGGCGCCGTGGTTGTGGCTGCCGGCGGAGCCCTGGCGGCGCGTTGGGAGGCAGTGCGCAGGCGCAGAAGGAGGAGGCGCGAGGACGTAGTGCTCACCCCATTGCACTTTACCTTTCCCGCCGACGAGGCACGACGCGTCGGTGAGGGTATGGAGACTATGCTGTCCTGCTGGCTTCAGCAACTTCATGAATTCGGAGGCCCCGAACTGGAGCGACCAGACCTTCTAAAGAGACCTGGGGTCACAGCACGAGTGCCTTCTGCTCCCTCTTCCACTTGCAGTGTCAACCGTGTTGTGGTAGATAGACGAATAAGATATAAGGGCGATGCGGTGCACTTGAGGCACTTGCAAGTGACAGGTACATTGGAGCTCAAACGCAAATCGACAGATGTACTTCTATTGATGCAAAACTTGCGACAGGAAAATGTCAATCCTTTCATAGGTTGTCTCTGCGAAGGTCGACCGGCGCTGGTGTTCGACGAGTGCTGGCGCGGCTCGCTGGAGGACGTGCTGATGGCGGACGACATCAAGCTGGACTGGACGTTCCGGCTGTCGCTGCTGTCGGACCTGGTGAAGGGCATGCGCTACCTGCACGGCTCCCCGCTGCGCGCGCACGGCCGCCTCTCCTCGCGGAACTGCGTGGTGGACTCGCGCTGGGTGCTGCGCGTCACCGACTACGGGCTGCCGGCGTTGCACCGCACGCAGGGACTGCCGCCCGCGCAGCGCACGCCCAGAGAACTTCTGTGGACGGCTCCCGAACTGATACGGGAGGCCCGAGAGGGCGGGGGCTGGCTCGCCACGCAGTCCGGGGACGTGTTCTCGTTCGCCATCATCATGCAGGAGGTCATAGTGCGCGGCGAGCCGTACTGCATGCTCTCCTTCACGCCTGAAGAGATCATCGACAAGCTGTGTCGTCCTCCGCCGTTGATTCGTCCGTCGGTGTCGATGGGCGCGGCGCCGCCCGAGGCCGTGAGCGTGATGCGGCAGTGCTGGAGCGAGGCGCCCGACCTGCGCCCCGACTTCAACCGCCTGCACGACGTGTTCCGGCAGATGCAGCGCGGCCGAAAAATTAATATTGTCGACTCCATGTTCGAAATGCTCGAGAAGTATAGCAATAATTTGGAAGAATTGATCAGGGAACGTACAGAGCAACTGGACATGGAAAAGAAAAAGACCGAACAACTACTTAATAGGATGCTGCCGAGATCAGTCGCGGAAAGACTAATGTTAGGATCGCGAGTCGAGCCTGAGGAGTTCGAAGAAGTGTCTATATACTTCAGCGACATTGTGGGATTCACGGCACTCGCAGCAAGGTCTACAGCGGTGCAGGTGGTGGACCTGCTCAACGACCTGTACACGACCTTCGACGCGGCCATCGAGCAGTACCGCGTCTACAAGGTGGAGACCATCGGCGACGCCTACATGGTAGTGGGCGGACTGCCGCAGCGCACCCGCGACCATGCCGAGAGCGTGGCCACCATGGCGCTGCACCTGCTGCACCTGGCGGGGCGCTTCCGCGTGCGGCACCTGCCCGACACCCCCCTGCACCTGCGCATCGGCCTGCACACGGGGCCCTGCTGCGCCGGGGTCGTCGGCCTCACCATGCCGCGCTACTGCCTCTTCGGAGACACCGTCAACACCGCCTCTAGGATGGAATCTACAGGAGCAGCTTGGCGTATCCAAATATCCAGTACAACAGCTGAGAAGCTACTGGCTGCGGGCGGGTACAAGCTACGATCCAGAGGGCTCACTCAGATCAAAGGCAAGGGTGCCATGCACACGTACTGGCTGCTTGGTAAGGAGGGATTCGATCCTCCGCTTCCGACCCCTCCACCGCTTGATTGCGAAGGTGAGGTCCTGCTGGAGGTGGAGGGAGAGAGCGAGCCTGAAGCGGCTCCCGTGGACTCCGGCCTCTCCTCCGCGTCGCCGGACAACAGGCCAGCTCCCTATCTCACAAGTTCAGAGCAGTCTCGTCGGCCGGTGTCGAGCGCGACCTCCGCGGACAGCTGCTCGCCGCCGCCGGAGCCACGAGGCTCACGAGCCCTGCTGCCCACCACCAGCGTCGACCACAGCGCGCCCTACGCACGATATAGATGGCTGGGCGGCAACGCACGTACTCTGAAGCGGCAGTGGTCCCTGGAGCGCGGCGACGCGCTGGCGGCGGCGGCGGCGAGCGACGCGCCAGCCGGCAGTCGCGTCACGCCGCGCCACGCGCCGCGCTACCGGACGCGCGCCGAGCCCGCCGACGACGACTAG
Protein
MRALPAVLTTLTALTVLTVLTTLTALSVHAHAGPPALLVLRSACFRQDLPRLHPIPRATLLSHQETCDVGWNERVTRRAVSTLTDGGLVEIIGWPGAAVCESISVTASVLRRPQPASCLRLKAQAAAVAELARRLRWSRALLPRAPPSSDCEAALRELTLFLLETGLLVRRAMLSARAFADRPEVLILCVSVAAKIGSKIESMLKDTDVVILLLHFEELMRDATDFANKDIENFKTMKSESIKASKVNYSSDSTSEREINDHKLNHEMVQNLNSPLSFYNIHRKMQIRSGKSSMNSTELSTNQLIVPFTKPSEGNQTEPANIVHEGTRSLDDRRDARGTTMSTKHAEIESSQTLQISKKDLRYLEIFSHFNDLFPGWLLILSENIFVEEHLTSDSISEKVTKVLESVEESERARQQSVQRRFLLYVRSGDRSAWSRAATLATNLTRALDGDRPWAATVALHDWHALERWNGTKPEDERDHWNGSNILATVLGAGGAVVVAAGGALAARWEAVRRRRRRRREDVVLTPLHFTFPADEARRVGEGMETMLSCWLQQLHEFGGPELERPDLLKRPGVTARVPSAPSSTCSVNRVVVDRRIRYKGDAVHLRHLQVTGTLELKRKSTDVLLLMQNLRQENVNPFIGCLCEGRPALVFDECWRGSLEDVLMADDIKLDWTFRLSLLSDLVKGMRYLHGSPLRAHGRLSSRNCVVDSRWVLRVTDYGLPALHRTQGLPPAQRTPRELLWTAPELIREAREGGGWLATQSGDVFSFAIIMQEVIVRGEPYCMLSFTPEEIIDKLCRPPPLIRPSVSMGAAPPEAVSVMRQCWSEAPDLRPDFNRLHDVFRQMQRGRKINIVDSMFEMLEKYSNNLEELIRERTEQLDMEKKKTEQLLNRMLPRSVAERLMLGSRVEPEEFEEVSIYFSDIVGFTALAARSTAVQVVDLLNDLYTTFDAAIEQYRVYKVETIGDAYMVVGGLPQRTRDHAESVATMALHLLHLAGRFRVRHLPDTPLHLRIGLHTGPCCAGVVGLTMPRYCLFGDTVNTASRMESTGAAWRIQISSTTAEKLLAAGGYKLRSRGLTQIKGKGAMHTYWLLGKEGFDPPLPTPPPLDCEGEVLLEVEGESEPEAAPVDSGLSSASPDNRPAPYLTSSEQSRRPVSSATSADSCSPPPEPRGSRALLPTTSVDHSAPYARYRWLGGNARTLKRQWSLERGDALAAAAASDAPAGSRVTPRHAPRYRTRAEPADDD
Summary
Catalytic Activity
GTP = 3',5'-cyclic GMP + diphosphate
Similarity
Belongs to the adenylyl cyclase class-4/guanylyl cyclase family.
Feature
chain Guanylate cyclase
Uniprot
EC Number
4.6.1.2
Pubmed
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
PDB
3ET6
E-value=2.47142e-41,
Score=429
Ontologies
PATHWAY
GO
Topology
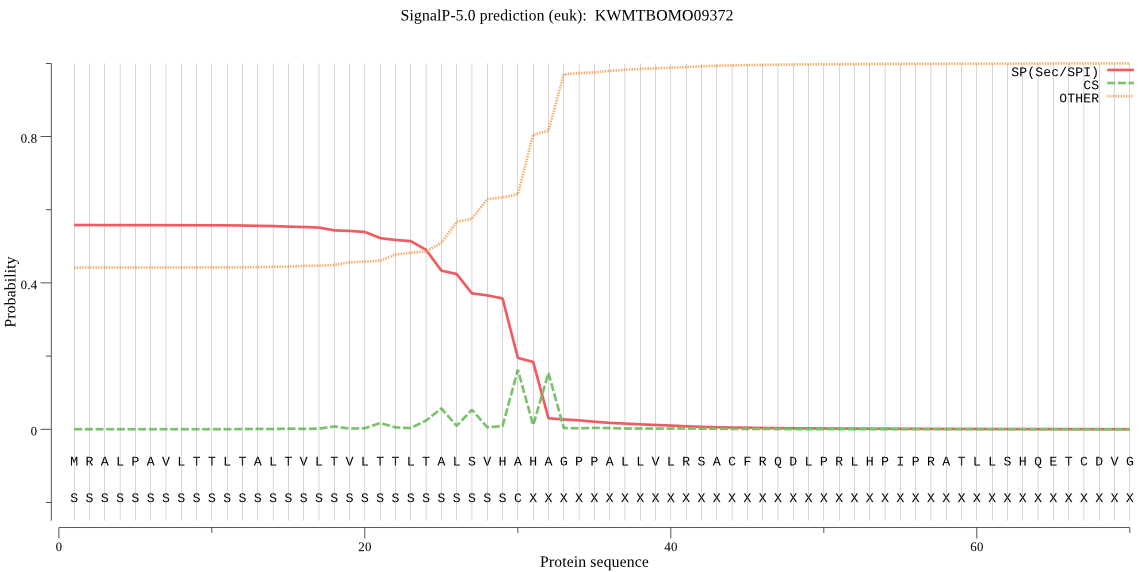
SignalP
Position: 1 - 30,
Likelihood: 0.557909
Length:
1244
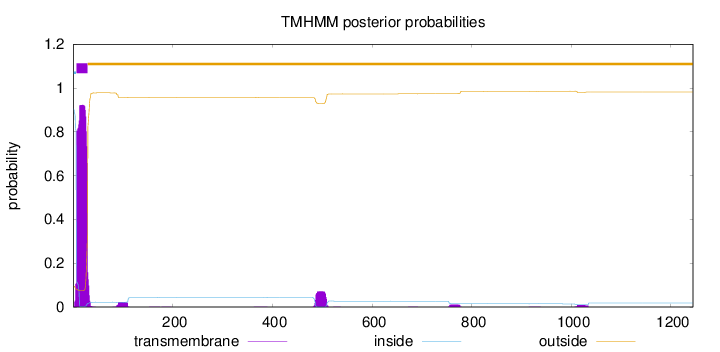
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.3022599999999
Exp number, first 60 AAs:
20.71583
Total prob of N-in:
0.90523
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 1244
Population Genetic Test Statistics
Pi
200.908558
Theta
180.894621
Tajima's D
0.617803
CLR
101.036896
CSRT
0.544022798860057
Interpretation
Possibly Positive selection
