Gene
KWMTBOMO09365
Pre Gene Modal
BGIBMGA002190
Annotation
PREDICTED:_diamine_acetyltransferase_2-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 3.418
Sequence
CDS
ATGTCTAACAAATATAAAAGCGGTGAAGTTATAGTCCGTGCAGCAACTAAACATGATATGAGTGCAGTCGCTGAAATGATACAGGAATTAGCAGATTACGAGAAAATGTCTGATGGTCCAAAATTATCCATTAAAGATTTGGTGCATGACGGCTTTGAAAAACAACCACCTGAGTTCTTCTGTAAAGTTGCAGAAAAACACAAAGAAAATATCTTAGGTTATGCTATTTACTTTCCTGTGTATTCCACGTGGGAGGGACGTGCGCTAATGCTGGAGGATCTGTACGTGAGAATGAATGAGCGGCGCAGAGGTGTTGGTAGATTGCTTTTCGATGCAGTTGCAAAGGAAGCGTCAGCGACGGGCTGCTGCCGCCTGGACTTCCACGTCCTGGAGTGGAATCCTGCTTGTTCGTTTTACGAGAGCAAAGGTGCGGTAAATCTAACTAACAAGGAGCAGTGGTGCTACTACAGGCTTACGGGAGATGCATTACGTGACTTTGCCAAAGATGGCTAG
Protein
MSNKYKSGEVIVRAATKHDMSAVAEMIQELADYEKMSDGPKLSIKDLVHDGFEKQPPEFFCKVAEKHKENILGYAIYFPVYSTWEGRALMLEDLYVRMNERRRGVGRLLFDAVAKEASATGCCRLDFHVLEWNPACSFYESKGAVNLTNKEQWCYYRLTGDALRDFAKDG
Summary
Uniprot
A0A2H1WBC0
A0A3S2LWV3
S4P0I9
A0A212FKE3
A0A0N1PFW3
A0A194Q1F4
+ More
A0A0L7KUQ5 A0A1B6LZY8 A0A0J7KFN4 A0A2A3EDH4 A0A026VTM7 E2AM98 A0A444T9P4 D6WL85 A0A0M3QYI6 A0A151WY43 B4LY24 B4KAG3 A0A194Q0P1 A0A195CVB1 B4NBB2 F4WCS1 A0A088AS02 A0A195DF76 A0A195ETW0 A0A195BVX8 B4JGB0 B4HE10 Q8MRT7 A0A1L8EDF7 A0A0A1WD58 B4QZ43 A0A1I8P0Q0 J3JUD9 A0A0K8UUY0 A0A034WLI5 B3MXA3 A0A1B6BXJ0 A0A1J1IIR2 B3P0R6 A0A1L8DC54 A0A158NQ88 E9GWR7 A0A1A9UYI1 A0A0P5REY2 B4PS81 A0A195CV71 Q294U0 A0A1W4W939 A0A162QMU6 A0A0P6B7J5 A0A0N1IC82 B4GMM0 B5DUI5 A0A1B0BBM7 B0WLN2 A0A154PGU4 W8BEZ6 A0A1L8ED03 A0A1A9YKZ9 A0A195CV67 A0A1B6EL94 A0A1B0A0F1 A0A3B0JN81 A0A182NMQ1 A0A232F6G7 K7IUH5 A0A1B6K3T3 A0A195ET61 A0A0L0CAF8 A0A195CVL0 A0A1A9W2C4 E2BV25 A0A1W4Y0H3 A0A182Q4L8 A0A1B0CI92 A0A3P8ZP28 A0A3B4U9C7 A0A3B4Y246 A0A1I8NHJ4 A0A1W4W0E7 F7EGI7 F4WCS0 A0A2W1BUP1 A0A195DGG0 A3KNF6 A0A3B3ZGL7 A0A3Q2YLV6 A0A1A8M6N9 A0A1A8FQD1 A0A0V0GA53 A0A3Q3MR83 A0A069DNJ1 A0A023F8G4 A0A1B0GF52 A0A0F8AJV2 A0A1A8U9V2 A0A3B1INP7 A0A3Q1IRT9
A0A0L7KUQ5 A0A1B6LZY8 A0A0J7KFN4 A0A2A3EDH4 A0A026VTM7 E2AM98 A0A444T9P4 D6WL85 A0A0M3QYI6 A0A151WY43 B4LY24 B4KAG3 A0A194Q0P1 A0A195CVB1 B4NBB2 F4WCS1 A0A088AS02 A0A195DF76 A0A195ETW0 A0A195BVX8 B4JGB0 B4HE10 Q8MRT7 A0A1L8EDF7 A0A0A1WD58 B4QZ43 A0A1I8P0Q0 J3JUD9 A0A0K8UUY0 A0A034WLI5 B3MXA3 A0A1B6BXJ0 A0A1J1IIR2 B3P0R6 A0A1L8DC54 A0A158NQ88 E9GWR7 A0A1A9UYI1 A0A0P5REY2 B4PS81 A0A195CV71 Q294U0 A0A1W4W939 A0A162QMU6 A0A0P6B7J5 A0A0N1IC82 B4GMM0 B5DUI5 A0A1B0BBM7 B0WLN2 A0A154PGU4 W8BEZ6 A0A1L8ED03 A0A1A9YKZ9 A0A195CV67 A0A1B6EL94 A0A1B0A0F1 A0A3B0JN81 A0A182NMQ1 A0A232F6G7 K7IUH5 A0A1B6K3T3 A0A195ET61 A0A0L0CAF8 A0A195CVL0 A0A1A9W2C4 E2BV25 A0A1W4Y0H3 A0A182Q4L8 A0A1B0CI92 A0A3P8ZP28 A0A3B4U9C7 A0A3B4Y246 A0A1I8NHJ4 A0A1W4W0E7 F7EGI7 F4WCS0 A0A2W1BUP1 A0A195DGG0 A3KNF6 A0A3B3ZGL7 A0A3Q2YLV6 A0A1A8M6N9 A0A1A8FQD1 A0A0V0GA53 A0A3Q3MR83 A0A069DNJ1 A0A023F8G4 A0A1B0GF52 A0A0F8AJV2 A0A1A8U9V2 A0A3B1INP7 A0A3Q1IRT9
Pubmed
23622113
22118469
26354079
26227816
24508170
30249741
+ More
20798317 18362917 19820115 17994087 21719571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 22516182 23537049 25348373 21347285 21292972 17550304 15632085 24495485 28648823 20075255 26108605 25069045 25315136 20431018 28756777 27762356 25463417 26334808 25474469 25835551 25329095
20798317 18362917 19820115 17994087 21719571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 22516182 23537049 25348373 21347285 21292972 17550304 15632085 24495485 28648823 20075255 26108605 25069045 25315136 20431018 28756777 27762356 25463417 26334808 25474469 25835551 25329095
EMBL
ODYU01007522
SOQ50371.1
RSAL01000141
RVE46073.1
GAIX01012920
JAA79640.1
+ More
AGBW02008061 OWR54213.1 KQ461033 KPJ09984.1 KQ459581 KPI99153.1 JTDY01005441 KOB66997.1 GEBQ01010728 JAT29249.1 LBMM01008229 KMQ89036.1 KZ288271 PBC29750.1 KK107965 QOIP01000001 EZA47040.1 RLU27410.1 GL440775 EFN65440.1 SAUD01006978 RXG62373.1 KQ971343 EFA03491.1 CP012526 ALC47743.1 KQ982662 KYQ52615.1 CH940650 EDW66890.1 CH933806 EDW15676.1 KPI99152.1 KQ977259 KYN04570.1 CH964232 EDW81076.2 GL888070 EGI68251.1 KQ980905 KYN11555.1 KQ981979 KYN31347.1 KQ976398 KYM92737.1 CH916369 EDV92579.1 CH480815 EDW42102.1 AE014297 AY119275 AAF55142.2 AAM51135.1 GFDG01002190 JAV16609.1 GBXI01017373 JAC96918.1 CM000364 EDX12871.1 APGK01040619 APGK01040620 APGK01040621 BT126852 KB740984 KB630727 KB632357 AEE61814.1 ENN76190.1 ERL83819.1 ERL93443.1 GDHF01022159 GDHF01014301 GDHF01012051 JAI30155.1 JAI38013.1 JAI40263.1 GAKP01004319 JAC54633.1 CH902629 EDV35326.1 GEDC01031296 JAS06002.1 CVRI01000047 CRK98345.1 CH954181 EDV48964.1 GFDF01010036 JAV04048.1 ADTU01001692 GL732571 EFX75947.1 GDIQ01103512 JAL48214.1 CM000160 EDW97501.1 KYN04571.1 CM000070 EAL28874.1 LRGB01000311 KZS19809.1 GDIP01032960 JAM70755.1 KPJ09983.1 CH479185 EDW38094.1 CH673789 EDY71551.1 JXJN01011542 DS231988 EDS30574.1 KQ434892 KZC10538.1 GAMC01014736 GAMC01014734 GAMC01014732 JAB91821.1 GFDG01002188 JAV16611.1 KYN04573.1 GECZ01031072 JAS38697.1 OUUW01000008 SPP83687.1 NNAY01000861 OXU26172.1 AAZX01002659 GECU01023370 GECU01001585 JAS84336.1 JAT06122.1 KYN31346.1 JRES01000776 KNC28424.1 KYN04572.1 GL450787 EFN80456.1 AXCN02001211 AJWK01013106 AAMC01148488 EGI68250.1 KZ149955 PZC76500.1 KYN11554.1 BC133814 CM004470 AAI33815.1 OCT90971.1 HAEF01011245 SBR51929.1 HAEB01015068 HAEC01001601 SBQ61595.1 GECL01001185 JAP04939.1 GBGD01003251 JAC85638.1 GBBI01001097 JAC17615.1 CCAG010012761 KQ041944 KKF20565.1 HAEJ01004438 SBS44895.1
AGBW02008061 OWR54213.1 KQ461033 KPJ09984.1 KQ459581 KPI99153.1 JTDY01005441 KOB66997.1 GEBQ01010728 JAT29249.1 LBMM01008229 KMQ89036.1 KZ288271 PBC29750.1 KK107965 QOIP01000001 EZA47040.1 RLU27410.1 GL440775 EFN65440.1 SAUD01006978 RXG62373.1 KQ971343 EFA03491.1 CP012526 ALC47743.1 KQ982662 KYQ52615.1 CH940650 EDW66890.1 CH933806 EDW15676.1 KPI99152.1 KQ977259 KYN04570.1 CH964232 EDW81076.2 GL888070 EGI68251.1 KQ980905 KYN11555.1 KQ981979 KYN31347.1 KQ976398 KYM92737.1 CH916369 EDV92579.1 CH480815 EDW42102.1 AE014297 AY119275 AAF55142.2 AAM51135.1 GFDG01002190 JAV16609.1 GBXI01017373 JAC96918.1 CM000364 EDX12871.1 APGK01040619 APGK01040620 APGK01040621 BT126852 KB740984 KB630727 KB632357 AEE61814.1 ENN76190.1 ERL83819.1 ERL93443.1 GDHF01022159 GDHF01014301 GDHF01012051 JAI30155.1 JAI38013.1 JAI40263.1 GAKP01004319 JAC54633.1 CH902629 EDV35326.1 GEDC01031296 JAS06002.1 CVRI01000047 CRK98345.1 CH954181 EDV48964.1 GFDF01010036 JAV04048.1 ADTU01001692 GL732571 EFX75947.1 GDIQ01103512 JAL48214.1 CM000160 EDW97501.1 KYN04571.1 CM000070 EAL28874.1 LRGB01000311 KZS19809.1 GDIP01032960 JAM70755.1 KPJ09983.1 CH479185 EDW38094.1 CH673789 EDY71551.1 JXJN01011542 DS231988 EDS30574.1 KQ434892 KZC10538.1 GAMC01014736 GAMC01014734 GAMC01014732 JAB91821.1 GFDG01002188 JAV16611.1 KYN04573.1 GECZ01031072 JAS38697.1 OUUW01000008 SPP83687.1 NNAY01000861 OXU26172.1 AAZX01002659 GECU01023370 GECU01001585 JAS84336.1 JAT06122.1 KYN31346.1 JRES01000776 KNC28424.1 KYN04572.1 GL450787 EFN80456.1 AXCN02001211 AJWK01013106 AAMC01148488 EGI68250.1 KZ149955 PZC76500.1 KYN11554.1 BC133814 CM004470 AAI33815.1 OCT90971.1 HAEF01011245 SBR51929.1 HAEB01015068 HAEC01001601 SBQ61595.1 GECL01001185 JAP04939.1 GBGD01003251 JAC85638.1 GBBI01001097 JAC17615.1 CCAG010012761 KQ041944 KKF20565.1 HAEJ01004438 SBS44895.1
Proteomes
UP000283053
UP000007151
UP000053240
UP000053268
UP000037510
UP000036403
+ More
UP000242457 UP000053097 UP000279307 UP000000311 UP000288706 UP000007266 UP000092553 UP000075809 UP000008792 UP000009192 UP000078542 UP000007798 UP000007755 UP000005203 UP000078492 UP000078541 UP000078540 UP000001070 UP000001292 UP000000803 UP000000304 UP000095300 UP000019118 UP000030742 UP000007801 UP000183832 UP000008711 UP000005205 UP000000305 UP000078200 UP000002282 UP000001819 UP000192223 UP000076858 UP000008744 UP000092460 UP000002320 UP000076502 UP000092443 UP000092445 UP000268350 UP000075884 UP000215335 UP000002358 UP000037069 UP000091820 UP000008237 UP000192224 UP000075886 UP000092461 UP000265140 UP000261420 UP000261360 UP000095301 UP000192221 UP000008143 UP000186698 UP000261520 UP000264820 UP000261640 UP000092444 UP000018467 UP000265040
UP000242457 UP000053097 UP000279307 UP000000311 UP000288706 UP000007266 UP000092553 UP000075809 UP000008792 UP000009192 UP000078542 UP000007798 UP000007755 UP000005203 UP000078492 UP000078541 UP000078540 UP000001070 UP000001292 UP000000803 UP000000304 UP000095300 UP000019118 UP000030742 UP000007801 UP000183832 UP000008711 UP000005205 UP000000305 UP000078200 UP000002282 UP000001819 UP000192223 UP000076858 UP000008744 UP000092460 UP000002320 UP000076502 UP000092443 UP000092445 UP000268350 UP000075884 UP000215335 UP000002358 UP000037069 UP000091820 UP000008237 UP000192224 UP000075886 UP000092461 UP000265140 UP000261420 UP000261360 UP000095301 UP000192221 UP000008143 UP000186698 UP000261520 UP000264820 UP000261640 UP000092444 UP000018467 UP000265040
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A2H1WBC0
A0A3S2LWV3
S4P0I9
A0A212FKE3
A0A0N1PFW3
A0A194Q1F4
+ More
A0A0L7KUQ5 A0A1B6LZY8 A0A0J7KFN4 A0A2A3EDH4 A0A026VTM7 E2AM98 A0A444T9P4 D6WL85 A0A0M3QYI6 A0A151WY43 B4LY24 B4KAG3 A0A194Q0P1 A0A195CVB1 B4NBB2 F4WCS1 A0A088AS02 A0A195DF76 A0A195ETW0 A0A195BVX8 B4JGB0 B4HE10 Q8MRT7 A0A1L8EDF7 A0A0A1WD58 B4QZ43 A0A1I8P0Q0 J3JUD9 A0A0K8UUY0 A0A034WLI5 B3MXA3 A0A1B6BXJ0 A0A1J1IIR2 B3P0R6 A0A1L8DC54 A0A158NQ88 E9GWR7 A0A1A9UYI1 A0A0P5REY2 B4PS81 A0A195CV71 Q294U0 A0A1W4W939 A0A162QMU6 A0A0P6B7J5 A0A0N1IC82 B4GMM0 B5DUI5 A0A1B0BBM7 B0WLN2 A0A154PGU4 W8BEZ6 A0A1L8ED03 A0A1A9YKZ9 A0A195CV67 A0A1B6EL94 A0A1B0A0F1 A0A3B0JN81 A0A182NMQ1 A0A232F6G7 K7IUH5 A0A1B6K3T3 A0A195ET61 A0A0L0CAF8 A0A195CVL0 A0A1A9W2C4 E2BV25 A0A1W4Y0H3 A0A182Q4L8 A0A1B0CI92 A0A3P8ZP28 A0A3B4U9C7 A0A3B4Y246 A0A1I8NHJ4 A0A1W4W0E7 F7EGI7 F4WCS0 A0A2W1BUP1 A0A195DGG0 A3KNF6 A0A3B3ZGL7 A0A3Q2YLV6 A0A1A8M6N9 A0A1A8FQD1 A0A0V0GA53 A0A3Q3MR83 A0A069DNJ1 A0A023F8G4 A0A1B0GF52 A0A0F8AJV2 A0A1A8U9V2 A0A3B1INP7 A0A3Q1IRT9
A0A0L7KUQ5 A0A1B6LZY8 A0A0J7KFN4 A0A2A3EDH4 A0A026VTM7 E2AM98 A0A444T9P4 D6WL85 A0A0M3QYI6 A0A151WY43 B4LY24 B4KAG3 A0A194Q0P1 A0A195CVB1 B4NBB2 F4WCS1 A0A088AS02 A0A195DF76 A0A195ETW0 A0A195BVX8 B4JGB0 B4HE10 Q8MRT7 A0A1L8EDF7 A0A0A1WD58 B4QZ43 A0A1I8P0Q0 J3JUD9 A0A0K8UUY0 A0A034WLI5 B3MXA3 A0A1B6BXJ0 A0A1J1IIR2 B3P0R6 A0A1L8DC54 A0A158NQ88 E9GWR7 A0A1A9UYI1 A0A0P5REY2 B4PS81 A0A195CV71 Q294U0 A0A1W4W939 A0A162QMU6 A0A0P6B7J5 A0A0N1IC82 B4GMM0 B5DUI5 A0A1B0BBM7 B0WLN2 A0A154PGU4 W8BEZ6 A0A1L8ED03 A0A1A9YKZ9 A0A195CV67 A0A1B6EL94 A0A1B0A0F1 A0A3B0JN81 A0A182NMQ1 A0A232F6G7 K7IUH5 A0A1B6K3T3 A0A195ET61 A0A0L0CAF8 A0A195CVL0 A0A1A9W2C4 E2BV25 A0A1W4Y0H3 A0A182Q4L8 A0A1B0CI92 A0A3P8ZP28 A0A3B4U9C7 A0A3B4Y246 A0A1I8NHJ4 A0A1W4W0E7 F7EGI7 F4WCS0 A0A2W1BUP1 A0A195DGG0 A3KNF6 A0A3B3ZGL7 A0A3Q2YLV6 A0A1A8M6N9 A0A1A8FQD1 A0A0V0GA53 A0A3Q3MR83 A0A069DNJ1 A0A023F8G4 A0A1B0GF52 A0A0F8AJV2 A0A1A8U9V2 A0A3B1INP7 A0A3Q1IRT9
PDB
2Q4V
E-value=2.11592e-23,
Score=265
Ontologies
GO
PANTHER
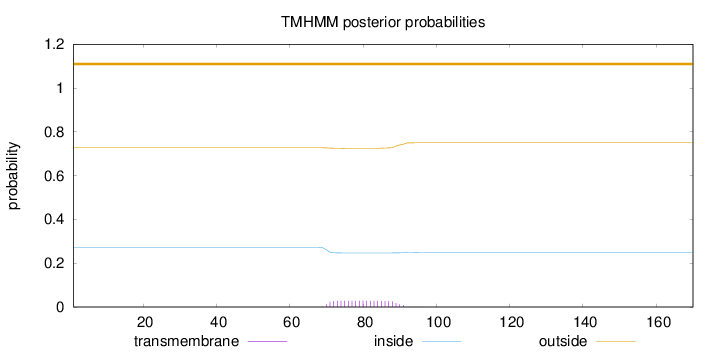
Topology
Length:
170
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.553939999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.27245
outside
1 - 170
Population Genetic Test Statistics
Pi
216.817126
Theta
186.732484
Tajima's D
0.314015
CLR
1.483601
CSRT
0.457427128643568
Interpretation
Possibly Positive selection
