Pre Gene Modal
BGIBMGA002162
Annotation
PREDICTED:_protocadherin-16_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 1.24
Sequence
CDS
ATGAATGGCGGCGCCGTGGAGAGCTTCTTCATCAGCGAGGACACACCCATCGGCAGTATTATAGGTACCTTAAGTGTAAATGGAGATCCTTCAGAAGACCTTGGAGACATCACCCTTCGGCTTCAAGAAAAGGGAGCAGCAGTAGGCATAGTGCCTGGTAGCAAGAACTTGACACTGCTCAGAGCTTTGGATCGAGAGGAGAAAATCGGCCCTTCTAATGTATACGTTAATGTACGGTGTGACAGAAGACGAACTACTGATCCGAGCTTCGTGATCCCGGTGTCGGTGCGCGTGTGGGACGTTAACGACAACGCGCCGCAGTGGGAGGGCGCGCCGTACGCCGTGCGGGTGTCGGAGACGGCGGCGAGGGGCGCGCGGGCGGCGGCCGTGCGCGCCTCCGACCGCGACCAGCCCGGCCCGCACTCCGCGCTGCACTACGCCGTGCTGCCCGGACCGCATTCCGAATATTTAGGTTTTGAAAATGAACTAGAACCAATAATAGTAGTTAAGAAGCCTCTGGACTTTGAGACAGTAAACAACTTCACAATAAAGCTCAGGGTACAAGACCAGGGCTCTCCTCCGCTGTACAGCGATACCACCCTGCAGGTCACAGTGCTGGATGCCGACGATCAGAACCCTAAGTTTACCTACGAACATTACACAGCTCTGCTGCCAGATGATGCTGAGGAGGGCACATTCCTCACGACCTCTCCCGGGCCGATATCAGCAGCCGACCAAGACTTGGGCATCAATGCTCCGCTGCAGTACTCCAGCACGGGAGACCACAAACAACTGATTGTCGTCAATAAGGACACTGGCCAGGTCGCCGCTACTAAACAGCTGATCGCAGAGTTGGATCCCTCCGTCACTATTGTTTTAAAAGCAACACAAGTGGATAATCCAGATCGGTACGCGTTAGCGACGCTTACCATCCGTAGTACCACACCACGATCTTCCACTCCGCCCATCAGCAGGCTTCAGTTCACCCAGAAGCAGTACTCGACCACAGTCAGTGAAGGAGCCTCGCCAGGATCCGTGCTCCTGACCCTAACCACCGTGCCACAAAATACAGAATTTAGGGAGAGTCCACTACAGTTCTACGTATCAGACCGCAGCTTCTTGGATAAATTTGCCATCAACAACGCCGGTGAAGTCATACTTAGGAGATCCCTTGACTATGAAACTACGTCCAATTACTTCTACCAAGTTATGGTCACTGACTCTATATCAAATGACACAGCATCAATAAACATAAGCGTGGAGGACGTGAATGAGTGGGAGCCGCGGTTCCGACACCCGCAGTACGAGTTCGAGGTGGAGGCGGCGGGGGGCGGGGCGGGGGAGGCGGCGGGGGGGCTGCGCGTGGGCCGGCTGGTGGCGCACGACGGGGACGCCGGCACGCTCGCGCTCAGCCTCGCCGGGCAGCACGCGCGTATGTTCTACATTAATGCCAGCGGAGATTTATTCCTGCGGGAGGAAGCGATCGGAAATTTAAACTCGTCATCTCTACACTTGGTGGCCACTGCTATAGATTCAGGGATTCCTCCACGACAGACATCGGTCCCGGTGACGGTGGTGGTGCGCGGGGCGGGGGGCGCGGCGGGGGGCGCGGTGCGGGGCGCGGGGCTGCTGGCGGCGCTGGTGGCGGCCCTCGCGCTGCTCGCGCTGCTCGTCTGTGGACTCTTCCTCTACATATACACCGCGAAAAAACGTATCAAATCTCCAGCACCGGCTCCACTCAAACCAGCTGGATTCATCAACCACGAGAAATCCCGACCGCTCCCCTCACCCCTGCCCCCCACCCTCCCCGACTCCGGCGCCAACGGCACCAACACTATTGCCAACAATGGCACAGCAACGCTGGCCACCAGCAGCTCCCTGCAGAGTGTCAGCGCCGGGGCTAGTACTATACTAGCCGGCTCTAGCTCCAGTCTTCATATGCCTGACGCTGCCACTAAGCCGTCAAATGGACACGAAGGTGCGAGGAGCGGGGAGGCGCGGCGGGGGGGCGGCGGGGGGGTGGCGCCGGCGGGCGGCGGGCGGGTGGCGCCGGCGGGCGGCGAGTGCGCCATGAGCGACCACCTGCGCGGCGGCGGCGGCGCGCGCTCCGGCGTGGCGTGGCCCGCGCACACCATCCCCGCCACGCTCAAGGCGCTCGCCTGGGACGACCACAACAAACAGAGCAGCGACGCAGAGAGTCCGGTGCACGACGCGACCAACCGCGCGGTCGCCGAGCACATGAACCTCACCGTGTACTTCTGA
Protein
MNGGAVESFFISEDTPIGSIIGTLSVNGDPSEDLGDITLRLQEKGAAVGIVPGSKNLTLLRALDREEKIGPSNVYVNVRCDRRRTTDPSFVIPVSVRVWDVNDNAPQWEGAPYAVRVSETAARGARAAAVRASDRDQPGPHSALHYAVLPGPHSEYLGFENELEPIIVVKKPLDFETVNNFTIKLRVQDQGSPPLYSDTTLQVTVLDADDQNPKFTYEHYTALLPDDAEEGTFLTTSPGPISAADQDLGINAPLQYSSTGDHKQLIVVNKDTGQVAATKQLIAELDPSVTIVLKATQVDNPDRYALATLTIRSTTPRSSTPPISRLQFTQKQYSTTVSEGASPGSVLLTLTTVPQNTEFRESPLQFYVSDRSFLDKFAINNAGEVILRRSLDYETTSNYFYQVMVTDSISNDTASINISVEDVNEWEPRFRHPQYEFEVEAAGGGAGEAAGGLRVGRLVAHDGDAGTLALSLAGQHARMFYINASGDLFLREEAIGNLNSSSLHLVATAIDSGIPPRQTSVPVTVVVRGAGGAAGGAVRGAGLLAALVAALALLALLVCGLFLYIYTAKKRIKSPAPAPLKPAGFINHEKSRPLPSPLPPTLPDSGANGTNTIANNGTATLATSSSLQSVSAGASTILAGSSSSLHMPDAATKPSNGHEGARSGEARRGGGGGVAPAGGGRVAPAGGECAMSDHLRGGGGARSGVAWPAHTIPATLKALAWDDHNKQSSDAESPVHDATNRAVAEHMNLTVYF
Summary
Uniprot
H9IY29
A0A437BY62
A0A194Q2C5
A0A212FKE2
A0A1B6IF97
A0A1B6EHK3
+ More
A0A2H8TDD0 A0A1I8N9G6 X1WJ38 A0A0L0BZZ6 A0A1A9Z6H2 B4KDJ1 A0A0A9Y7R0 A0A1I8PWZ1 A0A1A9V0C2 A0A1A9WGZ1 A0A0K8UD22 Q293R6 A0A0K8UDJ1 A0A0Q9WMI2 B4PUB2 A0A0M4EMZ3 A0A2S2R9D2 A0A0R3NK77 A0A3B0KDJ2 A0A1W4VMB2 B4JV09 A0A0Q9WF28 A0A1W4VKX6 B3LWI5 B4LZV0 A0A1A9YSK4 Q9VBV6 B4IJI5 A0A146KX84 A0A088ANY8 A0A0L7QN49 K7JAD0 A0A310SCB6 A0A154PNL4 A0A026WSE2 A0A0N0BFM1 A0A336MIQ2 A0A139WIG1 E0VWN8 A0A1B0DFZ1 A0A0T6AX74 A0A1Y1LMF3 E2C1G4 A0A1W4WGP4 A0A195C5Q2 A0A195ELT0 A0A195B5T3 Q172R3 A0A1S4FG21 A0A195ESQ3 A0A0K2TUI5 E2A6H9 A0A087TEE9 A0A1J1HUC9 A0A158NIT4 A0A0P5W7S9 A0A0P5TAJ2 A0A0P4X9N0 A0A182M724 A0A0P6JRU8 A0A2A3ERY1 B0W9C8 A0A182VXH8 A0A182REU1
A0A2H8TDD0 A0A1I8N9G6 X1WJ38 A0A0L0BZZ6 A0A1A9Z6H2 B4KDJ1 A0A0A9Y7R0 A0A1I8PWZ1 A0A1A9V0C2 A0A1A9WGZ1 A0A0K8UD22 Q293R6 A0A0K8UDJ1 A0A0Q9WMI2 B4PUB2 A0A0M4EMZ3 A0A2S2R9D2 A0A0R3NK77 A0A3B0KDJ2 A0A1W4VMB2 B4JV09 A0A0Q9WF28 A0A1W4VKX6 B3LWI5 B4LZV0 A0A1A9YSK4 Q9VBV6 B4IJI5 A0A146KX84 A0A088ANY8 A0A0L7QN49 K7JAD0 A0A310SCB6 A0A154PNL4 A0A026WSE2 A0A0N0BFM1 A0A336MIQ2 A0A139WIG1 E0VWN8 A0A1B0DFZ1 A0A0T6AX74 A0A1Y1LMF3 E2C1G4 A0A1W4WGP4 A0A195C5Q2 A0A195ELT0 A0A195B5T3 Q172R3 A0A1S4FG21 A0A195ESQ3 A0A0K2TUI5 E2A6H9 A0A087TEE9 A0A1J1HUC9 A0A158NIT4 A0A0P5W7S9 A0A0P5TAJ2 A0A0P4X9N0 A0A182M724 A0A0P6JRU8 A0A2A3ERY1 B0W9C8 A0A182VXH8 A0A182REU1
Pubmed
EMBL
BABH01029839
BABH01029840
RSAL01000001
RVE55344.1
KQ459581
KPI99149.1
+ More
AGBW02008061 OWR54215.1 GECU01022139 JAS85567.1 GECZ01032371 JAS37398.1 GFXV01000289 MBW12094.1 ABLF02033186 ABLF02033188 JRES01001097 KNC25561.1 CH933806 EDW13825.2 GBHO01015380 GDHC01003997 JAG28224.1 JAQ14632.1 GDHF01027775 JAI24539.1 CM000070 EAL29148.3 GDHF01027676 GDHF01023302 JAI24638.1 JAI29012.1 CH940650 KRF83143.1 CM000160 EDW98799.2 CP012526 ALC46902.1 GGMS01017392 MBY86595.1 KRT01319.1 OUUW01000008 SPP83806.1 CH916374 EDV91329.1 KRF83144.1 CH902617 EDV41579.2 EDW67178.1 AE014297 AAF56421.3 CH480848 EDW51176.1 GDHC01018394 JAQ00235.1 KQ414860 KOC60048.1 AAZX01000433 KQ770827 OAD52576.1 KQ435007 KZC13466.1 KK107119 QOIP01000009 EZA58576.1 RLU18593.1 KQ435796 KOX73538.1 UFQS01001064 UFQT01001064 SSX08801.1 SSX28713.1 KQ971342 KYB27694.1 DS235823 EEB17794.1 AJVK01059138 LJIG01022697 KRT79191.1 GEZM01053554 GEZM01053553 JAV74018.1 GL451937 EFN78140.1 KQ978292 KYM95511.1 KQ978739 KYN28882.1 KQ976582 KYM79846.1 CH477432 EAT41034.1 KQ981979 KYN31263.1 HACA01011705 CDW29066.1 GL437132 EFN70936.1 KK114845 KFM63488.1 CVRI01000021 CRK91613.1 ADTU01001832 ADTU01001833 ADTU01001834 GDIP01090088 JAM13627.1 GDIQ01094590 JAL57136.1 GDIP01244955 JAI78446.1 AXCM01000911 GDIQ01011065 JAN83672.1 KZ288193 PBC34032.1 DS231863 EDS39988.1
AGBW02008061 OWR54215.1 GECU01022139 JAS85567.1 GECZ01032371 JAS37398.1 GFXV01000289 MBW12094.1 ABLF02033186 ABLF02033188 JRES01001097 KNC25561.1 CH933806 EDW13825.2 GBHO01015380 GDHC01003997 JAG28224.1 JAQ14632.1 GDHF01027775 JAI24539.1 CM000070 EAL29148.3 GDHF01027676 GDHF01023302 JAI24638.1 JAI29012.1 CH940650 KRF83143.1 CM000160 EDW98799.2 CP012526 ALC46902.1 GGMS01017392 MBY86595.1 KRT01319.1 OUUW01000008 SPP83806.1 CH916374 EDV91329.1 KRF83144.1 CH902617 EDV41579.2 EDW67178.1 AE014297 AAF56421.3 CH480848 EDW51176.1 GDHC01018394 JAQ00235.1 KQ414860 KOC60048.1 AAZX01000433 KQ770827 OAD52576.1 KQ435007 KZC13466.1 KK107119 QOIP01000009 EZA58576.1 RLU18593.1 KQ435796 KOX73538.1 UFQS01001064 UFQT01001064 SSX08801.1 SSX28713.1 KQ971342 KYB27694.1 DS235823 EEB17794.1 AJVK01059138 LJIG01022697 KRT79191.1 GEZM01053554 GEZM01053553 JAV74018.1 GL451937 EFN78140.1 KQ978292 KYM95511.1 KQ978739 KYN28882.1 KQ976582 KYM79846.1 CH477432 EAT41034.1 KQ981979 KYN31263.1 HACA01011705 CDW29066.1 GL437132 EFN70936.1 KK114845 KFM63488.1 CVRI01000021 CRK91613.1 ADTU01001832 ADTU01001833 ADTU01001834 GDIP01090088 JAM13627.1 GDIQ01094590 JAL57136.1 GDIP01244955 JAI78446.1 AXCM01000911 GDIQ01011065 JAN83672.1 KZ288193 PBC34032.1 DS231863 EDS39988.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000007151
UP000095301
UP000007819
+ More
UP000037069 UP000092445 UP000009192 UP000095300 UP000078200 UP000091820 UP000001819 UP000008792 UP000002282 UP000092553 UP000268350 UP000192221 UP000001070 UP000007801 UP000092443 UP000000803 UP000001292 UP000005203 UP000053825 UP000002358 UP000076502 UP000053097 UP000279307 UP000053105 UP000007266 UP000009046 UP000092462 UP000008237 UP000192223 UP000078542 UP000078492 UP000078540 UP000008820 UP000078541 UP000000311 UP000054359 UP000183832 UP000005205 UP000075883 UP000242457 UP000002320 UP000075920 UP000075900
UP000037069 UP000092445 UP000009192 UP000095300 UP000078200 UP000091820 UP000001819 UP000008792 UP000002282 UP000092553 UP000268350 UP000192221 UP000001070 UP000007801 UP000092443 UP000000803 UP000001292 UP000005203 UP000053825 UP000002358 UP000076502 UP000053097 UP000279307 UP000053105 UP000007266 UP000009046 UP000092462 UP000008237 UP000192223 UP000078542 UP000078492 UP000078540 UP000008820 UP000078541 UP000000311 UP000054359 UP000183832 UP000005205 UP000075883 UP000242457 UP000002320 UP000075920 UP000075900
Interpro
Gene 3D
ProteinModelPortal
H9IY29
A0A437BY62
A0A194Q2C5
A0A212FKE2
A0A1B6IF97
A0A1B6EHK3
+ More
A0A2H8TDD0 A0A1I8N9G6 X1WJ38 A0A0L0BZZ6 A0A1A9Z6H2 B4KDJ1 A0A0A9Y7R0 A0A1I8PWZ1 A0A1A9V0C2 A0A1A9WGZ1 A0A0K8UD22 Q293R6 A0A0K8UDJ1 A0A0Q9WMI2 B4PUB2 A0A0M4EMZ3 A0A2S2R9D2 A0A0R3NK77 A0A3B0KDJ2 A0A1W4VMB2 B4JV09 A0A0Q9WF28 A0A1W4VKX6 B3LWI5 B4LZV0 A0A1A9YSK4 Q9VBV6 B4IJI5 A0A146KX84 A0A088ANY8 A0A0L7QN49 K7JAD0 A0A310SCB6 A0A154PNL4 A0A026WSE2 A0A0N0BFM1 A0A336MIQ2 A0A139WIG1 E0VWN8 A0A1B0DFZ1 A0A0T6AX74 A0A1Y1LMF3 E2C1G4 A0A1W4WGP4 A0A195C5Q2 A0A195ELT0 A0A195B5T3 Q172R3 A0A1S4FG21 A0A195ESQ3 A0A0K2TUI5 E2A6H9 A0A087TEE9 A0A1J1HUC9 A0A158NIT4 A0A0P5W7S9 A0A0P5TAJ2 A0A0P4X9N0 A0A182M724 A0A0P6JRU8 A0A2A3ERY1 B0W9C8 A0A182VXH8 A0A182REU1
A0A2H8TDD0 A0A1I8N9G6 X1WJ38 A0A0L0BZZ6 A0A1A9Z6H2 B4KDJ1 A0A0A9Y7R0 A0A1I8PWZ1 A0A1A9V0C2 A0A1A9WGZ1 A0A0K8UD22 Q293R6 A0A0K8UDJ1 A0A0Q9WMI2 B4PUB2 A0A0M4EMZ3 A0A2S2R9D2 A0A0R3NK77 A0A3B0KDJ2 A0A1W4VMB2 B4JV09 A0A0Q9WF28 A0A1W4VKX6 B3LWI5 B4LZV0 A0A1A9YSK4 Q9VBV6 B4IJI5 A0A146KX84 A0A088ANY8 A0A0L7QN49 K7JAD0 A0A310SCB6 A0A154PNL4 A0A026WSE2 A0A0N0BFM1 A0A336MIQ2 A0A139WIG1 E0VWN8 A0A1B0DFZ1 A0A0T6AX74 A0A1Y1LMF3 E2C1G4 A0A1W4WGP4 A0A195C5Q2 A0A195ELT0 A0A195B5T3 Q172R3 A0A1S4FG21 A0A195ESQ3 A0A0K2TUI5 E2A6H9 A0A087TEE9 A0A1J1HUC9 A0A158NIT4 A0A0P5W7S9 A0A0P5TAJ2 A0A0P4X9N0 A0A182M724 A0A0P6JRU8 A0A2A3ERY1 B0W9C8 A0A182VXH8 A0A182REU1
PDB
5DZV
E-value=1.60552e-19,
Score=239
Ontologies
GO
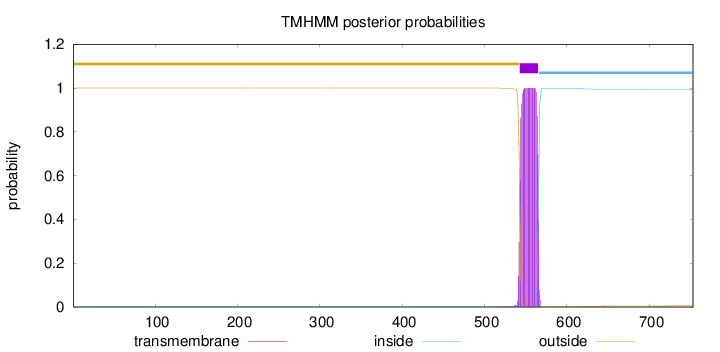
Topology
Length:
753
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.99984
Exp number, first 60 AAs:
0.00041
Total prob of N-in:
0.00026
outside
1 - 542
TMhelix
543 - 565
inside
566 - 753
Population Genetic Test Statistics
Pi
255.229704
Theta
197.530575
Tajima's D
0.943966
CLR
0.017574
CSRT
0.646117694115294
Interpretation
Possibly Positive selection
