Gene
KWMTBOMO09362
Pre Gene Modal
BGIBMGA002163
Annotation
hypothetical_protein_RR46_02365_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 3.28
Sequence
CDS
ATGCAATATCTAGTCCAACTCGTGGTGGCATTCCTCTTGACGACGCAGGCGAGTAGTGACAACAGCACGCTTTCAGAAGACACGTGCATCACGGAACACGGACTATGGGGCAGCCTGCTCGAAGAGTGGGCCGTCTCTGCAACTCCACAACATACAAGACAAGGCCGCATAATGGCGTTGCCCCCTGCCCACGGTTACTACGTATCTGACCGGATAGATCTCATGCCACCACCACCACCGCCTCCCCGCTTGTACCCCACTACTCAGCATCTGCCTCACCCCTCGCACTACAATGAGTGGCAGGCCAGTCCGCCACCGCCACCAGGACCTGGGAAGATTGTTAATCGACCACCAAATCCATATAAGGATAAATTTAAACCTAGTTATCCGCCTCCATCACAGAATCAGCCAATCCCAGTTCACACTCCGCAGCAGCCATACGCTCCTGGAGGATTAGTCGACCGGGTGGACGAGACCAAACCGAGCAAACCACAGAAGCAAGTTTCGGAAACCGATCTATATTTGCTAACCGCTATAGAGAAATTGGTTTATCGAGTGGACTTAATGGAGAAACGGTTGCGAAAAATGGAAGAAAACGTCCATTTCGTTTTGGCTGGAACTGATGTGAAGCCGGAGCCCTGCGCCAGTAACTTTACTCGCGTCGGAAGCGGCTGCTATTACTTCTCGACGGACGCGATCAACTGGAAGAATGCCAACTACGCGTGTCGGAAACTGAAAGGCAACTTGTTGGAGCTCGACGCGGACGACGAGAAGCGGCACTTGTTCGCCAACTTGCTCTCCGACAACAGACTGAAGGGTGCAGATTATTGGACGGGAGGTCTGAATCCAGGTCTTCTATGGATCTGGTCCCACTCCGCCAAGCCGGTGTCATCCAACAATACAAACACAAGCAGCTCGCACAGCATCGCGGGCGAGGGTCGGTGCTTGGCGCTGGTGCATGACCCGGCTCTCCACTCCTACCTGTACCGCGGACAGGACTGCGCTCTGAGACACAGATATGTTTGCGAAAAGGAAGAAGATAATGACAAGTTGGGGAATGAAATAGAAAGGGTCGCCAAGAAATTGAGAGAAGTTAAGAGGAAATCTAGAATTTTGATTAGTGGAGAAGACCCGTGA
Protein
MQYLVQLVVAFLLTTQASSDNSTLSEDTCITEHGLWGSLLEEWAVSATPQHTRQGRIMALPPAHGYYVSDRIDLMPPPPPPPRLYPTTQHLPHPSHYNEWQASPPPPPGPGKIVNRPPNPYKDKFKPSYPPPSQNQPIPVHTPQQPYAPGGLVDRVDETKPSKPQKQVSETDLYLLTAIEKLVYRVDLMEKRLRKMEENVHFVLAGTDVKPEPCASNFTRVGSGCYYFSTDAINWKNANYACRKLKGNLLELDADDEKRHLFANLLSDNRLKGADYWTGGLNPGLLWIWSHSAKPVSSNNTNTSSSHSIAGEGRCLALVHDPALHSYLYRGQDCALRHRYVCEKEEDNDKLGNEIERVAKKLREVKRKSRILISGEDP
Summary
Uniprot
EMBL
BABH01029837
NWSH01002071
PCG69249.1
KZ149955
PZC76488.1
RSAL01000001
+ More
RVE55345.1 AGBW02008061 OWR54217.1 APGK01037191 KB740948 ENN77256.1 JXUM01095358 KQ564191 KXJ72601.1 CVRI01000047 CRK97789.1 GAMC01005375 JAC01181.1 CH478095 EAT33985.1 CH916369 EDV92605.1 CH933806 EDW15155.2 JXJN01030917 CP012526 ALC47374.1
RVE55345.1 AGBW02008061 OWR54217.1 APGK01037191 KB740948 ENN77256.1 JXUM01095358 KQ564191 KXJ72601.1 CVRI01000047 CRK97789.1 GAMC01005375 JAC01181.1 CH478095 EAT33985.1 CH916369 EDV92605.1 CH933806 EDW15155.2 JXJN01030917 CP012526 ALC47374.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF56436
SSF56436
Gene 3D
ProteinModelPortal
PDB
3J82
E-value=0.000615007,
Score=102
Ontologies
Topology
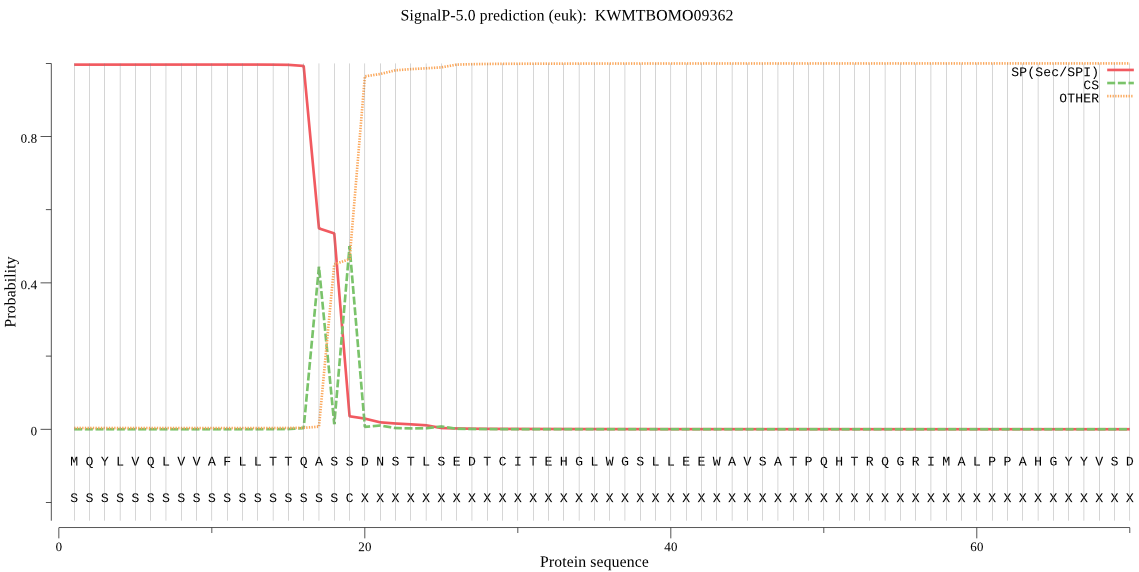
SignalP
Position: 1 - 19,
Likelihood: 0.996472
Length:
378
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00563
Exp number, first 60 AAs:
0.00264
Total prob of N-in:
0.00635
outside
1 - 378

Population Genetic Test Statistics
Pi
257.894636
Theta
157.086526
Tajima's D
-1.62926
CLR
1319.461117
CSRT
0.0451977401129944
Interpretation
Possibly Positive selection
