Gene
KWMTBOMO09355 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002186
Annotation
thiol_peroxiredoxin_[Bombyx_mori]
Full name
Peroxiredoxin 1
Alternative Name
Cytosolic thioredoxin peroxidase
Thioredoxin peroxidase
Thioredoxin peroxidase
Location in the cell
Cytoplasmic Reliability : 2.647
Sequence
CDS
ATGCCTCTGCAGATGACCAAACCCGCTCCCCAGTTCAAGGCCACGGCCGTCGTCAACGGAGAGTTCAAGGACATTTCTCTGTCTGACTACAAGGGGAAATATGTTGTGCTGTTCTTCTATCCTTTGGACTTCACGTTCGTGTGCCCGACGGAGATCATCGCGTTCTCGGAGAAGGCGGACGAGTTCCGCAAGATCGGCTGCGAGGTGCTCGGCGCCTCCACCGACTCGCACTTCACGCACCTCGCCTGGATCAACACGCCGCGCAAGCAGGGCGGACTCGGTCCCATGAACATCCCTCTGATAAGCGACAAGTCGCACCGCATCTCCCGCGACTACGGAGTGCTGGACGAGGAGACGGGCATCCCCTTCCGAGGACTCTTCATCATCGACGACAAGCAGAACCTCAGGCAGATCACCATCAACGACCTGCCCGTGGGGAGGTCGGTGGAGGAGACCCTGCGGCTGGTGCAGGCGTTCCAGTTCACGGACAAGCACGGCGAGGTGTGCCCCGCCAACTGGAGGCCCGGCGCCAAGACCATCAAGCCCGACACCAAGGCCGCGCAGGAGTACTTCGGCGACGCCAACTAG
Protein
MPLQMTKPAPQFKATAVVNGEFKDISLSDYKGKYVVLFFYPLDFTFVCPTEIIAFSEKADEFRKIGCEVLGASTDSHFTHLAWINTPRKQGGLGPMNIPLISDKSHRISRDYGVLDEETGIPFRGLFIIDDKQNLRQITINDLPVGRSVEETLRLVQAFQFTDKHGEVCPANWRPGAKTIKPDTKAAQEYFGDAN
Summary
Description
Thiol-specific peroxidase that catalyzes the reduction of hydrogen peroxide and organic hydroperoxides to water and alcohols, respectively. Plays a role in cell protection against oxidative stress by detoxifying peroxides and as sensor of hydrogen peroxide-mediated signaling events. Might participate in the signaling cascades of growth factors and tumor necrosis factor-alpha by regulating the intracellular concentrations of H(2)O(2) (By similarity). Reduces an intramolecular disulfide bond in GDPD5 that gates the ability to GDPD5 to drive postmitotic motor neuron differentiation (By similarity).
Catalytic Activity
[protein]-dithiol + a hydroperoxide = [protein]-disulfide + an alcohol + H2O
Subunit
Homodimer; disulfide-linked, upon oxidation. 5 homodimers assemble to form a ring-like decamer (By similarity). Interacts with GDPD5; forms a mixed-disulfide with GDPD5 (By similarity). Interacts with SESN1 and SESN2 (By similarity).
Miscellaneous
The active site is a conserved redox-active cysteine residue, the peroxidatic cysteine (C(P)), which makes the nucleophilic attack on the peroxide substrate. The peroxide oxidizes the C(P)-SH to cysteine sulfenic acid (C(P)-SOH), which then reacts with another cysteine residue, the resolving cysteine (C(R)), to form a disulfide bridge. The disulfide is subsequently reduced by an appropriate electron donor to complete the catalytic cycle. In this typical 2-Cys peroxiredoxin, C(R) is provided by the other dimeric subunit to form an intersubunit disulfide. The disulfide is subsequently reduced by thioredoxin (By similarity). As a reducing substrate, thioredoxin 2 is preferred over thioredoxin 1 (PubMed:11877442).
Similarity
Belongs to the peroxiredoxin family. AhpC/Prx1 subfamily.
Keywords
Antioxidant
Complete proteome
Cytoplasm
Disulfide bond
Oxidoreductase
Peroxidase
Phosphoprotein
Redox-active center
Reference proteome
Ubl conjugation
Feature
chain Peroxiredoxin 1
Uniprot
Q6T3A7
A0A088MGF5
A0A2S0RQQ1
A0A2L1CF95
A0A2A4IX67
A0A212EPQ2
+ More
B2KSE9 W0G2G4 A0A194QN77 A0A194Q0R0 I4DJ66 A0A3G1T1I6 A0A310SIG5 A0A0L7R4M0 A0A087ZND2 A0A154PDX5 E2BSB2 C3VVL4 A0A3L8DIR4 A0A026WMD6 A0A2A3E783 V9IJ36 T1P9R6 A0A0N0BCN5 A0A0L0C893 A0A1W4VJ93 A0A0M3QZ91 E2ALM7 B3MXP7 B4R4B3 B4IGR0 X2JF59 Q9V3P0 A0A067QT52 G0WRL1 A0A411G805 A0A195DBS0 Q6XJ14 B3NWP9 A0A151I178 A0A158NMR0 A0A3B0JT65 L0ATY2 A0A151X141 A0A151IBS9 A0A0R3P480 Q29IR1 B4H2V8 A0A2J7QKP8 A0A232ET67 A0A0C9QZH8 T1E316 B4NBT6 F4WVS6 A0A1L8EF12 A0A0Q9X143 A0A151JTE8 B4L6U4 A0A023EJ61 K7J7Q6 A0A2P8Z9A3 A0A0J7L7W7 A0A0C9QCP4 A0A0K8TWN4 A0A034W382 A0A1L7LQE0 B1N694 U5EZ75 W8BQT7 A0A1I8Q0P3 Q17DN4 Q8WSF6 V5K6U7 A2I476 A0A0K8TUD5 A0A3P8ZAF2 A0A0A1WZP3 B4MA19 A0A182T5T8 A0A0F6MVY1 A0A182JGY2 A0A0B6Z6H1 A0A1L8EEU6 A0A0B6Z732 A0A3P9AQB1 A0A0B6Z4Q3 A0A3N0YJY2 I3PU24 T1DN15 A0A146J914 A0A2I8B5Y1 A0A1B6F8I1 A0A1W4WZJ8 A0A1B6INE9 B5XBY3 A0A1D2AJ30 W5LIA3 C1KC72 B5X7X7 A0A0M3SH29
B2KSE9 W0G2G4 A0A194QN77 A0A194Q0R0 I4DJ66 A0A3G1T1I6 A0A310SIG5 A0A0L7R4M0 A0A087ZND2 A0A154PDX5 E2BSB2 C3VVL4 A0A3L8DIR4 A0A026WMD6 A0A2A3E783 V9IJ36 T1P9R6 A0A0N0BCN5 A0A0L0C893 A0A1W4VJ93 A0A0M3QZ91 E2ALM7 B3MXP7 B4R4B3 B4IGR0 X2JF59 Q9V3P0 A0A067QT52 G0WRL1 A0A411G805 A0A195DBS0 Q6XJ14 B3NWP9 A0A151I178 A0A158NMR0 A0A3B0JT65 L0ATY2 A0A151X141 A0A151IBS9 A0A0R3P480 Q29IR1 B4H2V8 A0A2J7QKP8 A0A232ET67 A0A0C9QZH8 T1E316 B4NBT6 F4WVS6 A0A1L8EF12 A0A0Q9X143 A0A151JTE8 B4L6U4 A0A023EJ61 K7J7Q6 A0A2P8Z9A3 A0A0J7L7W7 A0A0C9QCP4 A0A0K8TWN4 A0A034W382 A0A1L7LQE0 B1N694 U5EZ75 W8BQT7 A0A1I8Q0P3 Q17DN4 Q8WSF6 V5K6U7 A2I476 A0A0K8TUD5 A0A3P8ZAF2 A0A0A1WZP3 B4MA19 A0A182T5T8 A0A0F6MVY1 A0A182JGY2 A0A0B6Z6H1 A0A1L8EEU6 A0A0B6Z732 A0A3P9AQB1 A0A0B6Z4Q3 A0A3N0YJY2 I3PU24 T1DN15 A0A146J914 A0A2I8B5Y1 A0A1B6F8I1 A0A1W4WZJ8 A0A1B6INE9 B5XBY3 A0A1D2AJ30 W5LIA3 C1KC72 B5X7X7 A0A0M3SH29
EC Number
1.11.1.15
Pubmed
15607657
19121390
25191834
22118469
24296440
26354079
+ More
22651552 20798317 19808099 19932185 30249741 24508170 25315136 26108605 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 10632718 11677042 12537569 11877442 18327897 26715182 28953965 24845553 21382109 14525923 17550304 21347285 15632085 28648823 24330624 21719571 18057021 24945155 26483478 20075255 29403074 25348373 26760975 28076409 18226547 24495485 17510324 24420863 26369729 25069045 25830018 20433749 25329095 22450240
22651552 20798317 19808099 19932185 30249741 24508170 25315136 26108605 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 10632718 11677042 12537569 11877442 18327897 26715182 28953965 24845553 21382109 14525923 17550304 21347285 15632085 28648823 24330624 21719571 18057021 24945155 26483478 20075255 29403074 25348373 26760975 28076409 18226547 24495485 17510324 24420863 26369729 25069045 25830018 20433749 25329095 22450240
EMBL
BABH01029814
AY438331
AAR15420.1
KJ995804
AIN39487.1
MF979106
+ More
AWA45969.1 MF521978 AVC05622.1 NWSH01005971 PCG63782.1 AGBW02013414 OWR43468.1 EU131680 ABW96360.1 KC685523 KC685524 AHF48621.1 KQ461195 KPJ06809.1 KQ459581 KPI99141.1 AK401334 BAM17956.1 MG992413 AXY94851.1 KQ760761 OAD59133.1 KQ414657 KOC65769.1 KQ434885 KZC10041.1 GL450157 EFN81425.1 FJ895866 FJ895867 ACP44066.1 QOIP01000007 RLU20033.1 KK107152 EZA57227.1 KZ288357 PBC27142.1 JR047795 AEY60511.1 KA645439 AFP60068.1 KQ435897 KOX69212.1 JRES01000755 KNC28658.1 CP012528 ALC49010.1 ALC49013.1 GL440609 EFN65636.1 CH902630 EDV38512.1 CM000366 EDX17815.1 CH480836 EDW49010.1 AE014298 AHN59672.1 AHN59673.1 AHN59674.1 AF167098 AF321615 AF321616 AY070534 BT014630 KK853308 KDR08683.1 HM641254 HM641255 ADT65135.1 MH366217 QBB02026.1 KQ981010 KYN10321.1 AY231665 CM000162 AAR09688.1 EDX01561.1 CH954180 EDV47211.1 KQ976593 KYM79656.1 ADTU01020675 OUUW01000003 SPP78650.1 JX915909 AFZ78684.1 KQ982588 KYQ54117.1 KQ978078 KYM97181.1 CH379063 KRT06147.1 EAL32592.3 CH479204 EDW30675.1 NEVH01013260 PNF29153.1 NNAY01002315 OXU21560.1 GBYB01001115 GBYB01010548 JAG70882.1 JAG80315.1 GALA01000785 JAA94067.1 CH964239 EDW82295.1 GL888394 EGI61696.1 GFDG01001521 JAV17278.1 KRF99593.1 KQ981936 KYN32759.1 CH933812 EDW06090.1 KRG07143.1 JXUM01009055 GAPW01004617 KQ560275 JAC08981.1 KXJ83372.1 AAZX01009307 PYGN01000140 PSN53079.1 LBMM01000362 KMQ99389.1 GBYB01001084 JAG70851.1 GDHF01033829 GDHF01023805 GDHF01007797 JAI18485.1 JAI28509.1 JAI44517.1 GAKP01010201 GAKP01010200 JAC48752.1 FX983166 BAV93784.1 EF103377 ABO26635.1 GANO01001301 JAB58570.1 GAMC01002925 JAC03631.1 CH477292 EAT44541.2 AF323181 AAL37254.1 KC197034 AGU16435.1 EF070579 ABM55645.1 GDAI01000053 JAI17550.1 GBXI01013087 GBXI01010287 JAD01205.1 JAD04005.1 CH940655 EDW66078.1 JX021298 AGG68945.1 HACG01016621 CEK63486.1 GFDG01001542 JAV17257.1 HACG01016620 HACG01016622 CEK63485.1 CEK63487.1 HACG01016623 CEK63488.1 RJVU01037554 ROL46546.1 JF795590 AFG25786.2 GAMD01003255 JAA98335.1 LC049075 BAU79645.1 MF346707 AUT13197.1 GECZ01023546 JAS46223.1 GECU01019261 JAS88445.1 BT048552 BT057679 BT149987 ACI68353.1 ACM09551.1 AGH92591.1 GETE01000212 JAT79209.1 FJ812091 JX133230 MF319750 ACO36036.1 AFR11898.1 ASS34529.1 BT047146 ACI66947.1 KP192122 ALD62538.1
AWA45969.1 MF521978 AVC05622.1 NWSH01005971 PCG63782.1 AGBW02013414 OWR43468.1 EU131680 ABW96360.1 KC685523 KC685524 AHF48621.1 KQ461195 KPJ06809.1 KQ459581 KPI99141.1 AK401334 BAM17956.1 MG992413 AXY94851.1 KQ760761 OAD59133.1 KQ414657 KOC65769.1 KQ434885 KZC10041.1 GL450157 EFN81425.1 FJ895866 FJ895867 ACP44066.1 QOIP01000007 RLU20033.1 KK107152 EZA57227.1 KZ288357 PBC27142.1 JR047795 AEY60511.1 KA645439 AFP60068.1 KQ435897 KOX69212.1 JRES01000755 KNC28658.1 CP012528 ALC49010.1 ALC49013.1 GL440609 EFN65636.1 CH902630 EDV38512.1 CM000366 EDX17815.1 CH480836 EDW49010.1 AE014298 AHN59672.1 AHN59673.1 AHN59674.1 AF167098 AF321615 AF321616 AY070534 BT014630 KK853308 KDR08683.1 HM641254 HM641255 ADT65135.1 MH366217 QBB02026.1 KQ981010 KYN10321.1 AY231665 CM000162 AAR09688.1 EDX01561.1 CH954180 EDV47211.1 KQ976593 KYM79656.1 ADTU01020675 OUUW01000003 SPP78650.1 JX915909 AFZ78684.1 KQ982588 KYQ54117.1 KQ978078 KYM97181.1 CH379063 KRT06147.1 EAL32592.3 CH479204 EDW30675.1 NEVH01013260 PNF29153.1 NNAY01002315 OXU21560.1 GBYB01001115 GBYB01010548 JAG70882.1 JAG80315.1 GALA01000785 JAA94067.1 CH964239 EDW82295.1 GL888394 EGI61696.1 GFDG01001521 JAV17278.1 KRF99593.1 KQ981936 KYN32759.1 CH933812 EDW06090.1 KRG07143.1 JXUM01009055 GAPW01004617 KQ560275 JAC08981.1 KXJ83372.1 AAZX01009307 PYGN01000140 PSN53079.1 LBMM01000362 KMQ99389.1 GBYB01001084 JAG70851.1 GDHF01033829 GDHF01023805 GDHF01007797 JAI18485.1 JAI28509.1 JAI44517.1 GAKP01010201 GAKP01010200 JAC48752.1 FX983166 BAV93784.1 EF103377 ABO26635.1 GANO01001301 JAB58570.1 GAMC01002925 JAC03631.1 CH477292 EAT44541.2 AF323181 AAL37254.1 KC197034 AGU16435.1 EF070579 ABM55645.1 GDAI01000053 JAI17550.1 GBXI01013087 GBXI01010287 JAD01205.1 JAD04005.1 CH940655 EDW66078.1 JX021298 AGG68945.1 HACG01016621 CEK63486.1 GFDG01001542 JAV17257.1 HACG01016620 HACG01016622 CEK63485.1 CEK63487.1 HACG01016623 CEK63488.1 RJVU01037554 ROL46546.1 JF795590 AFG25786.2 GAMD01003255 JAA98335.1 LC049075 BAU79645.1 MF346707 AUT13197.1 GECZ01023546 JAS46223.1 GECU01019261 JAS88445.1 BT048552 BT057679 BT149987 ACI68353.1 ACM09551.1 AGH92591.1 GETE01000212 JAT79209.1 FJ812091 JX133230 MF319750 ACO36036.1 AFR11898.1 ASS34529.1 BT047146 ACI66947.1 KP192122 ALD62538.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000053825
+ More
UP000005203 UP000076502 UP000008237 UP000279307 UP000053097 UP000242457 UP000095301 UP000053105 UP000037069 UP000192221 UP000092553 UP000000311 UP000007801 UP000000304 UP000001292 UP000000803 UP000027135 UP000078492 UP000002282 UP000008711 UP000078540 UP000005205 UP000268350 UP000075809 UP000078542 UP000001819 UP000008744 UP000235965 UP000215335 UP000007798 UP000007755 UP000078541 UP000009192 UP000069940 UP000249989 UP000002358 UP000245037 UP000036403 UP000095300 UP000008820 UP000265140 UP000008792 UP000075901 UP000075880 UP000192223 UP000087266 UP000018467
UP000005203 UP000076502 UP000008237 UP000279307 UP000053097 UP000242457 UP000095301 UP000053105 UP000037069 UP000192221 UP000092553 UP000000311 UP000007801 UP000000304 UP000001292 UP000000803 UP000027135 UP000078492 UP000002282 UP000008711 UP000078540 UP000005205 UP000268350 UP000075809 UP000078542 UP000001819 UP000008744 UP000235965 UP000215335 UP000007798 UP000007755 UP000078541 UP000009192 UP000069940 UP000249989 UP000002358 UP000245037 UP000036403 UP000095300 UP000008820 UP000265140 UP000008792 UP000075901 UP000075880 UP000192223 UP000087266 UP000018467
Interpro
SUPFAM
SSF52833
SSF52833
ProteinModelPortal
Q6T3A7
A0A088MGF5
A0A2S0RQQ1
A0A2L1CF95
A0A2A4IX67
A0A212EPQ2
+ More
B2KSE9 W0G2G4 A0A194QN77 A0A194Q0R0 I4DJ66 A0A3G1T1I6 A0A310SIG5 A0A0L7R4M0 A0A087ZND2 A0A154PDX5 E2BSB2 C3VVL4 A0A3L8DIR4 A0A026WMD6 A0A2A3E783 V9IJ36 T1P9R6 A0A0N0BCN5 A0A0L0C893 A0A1W4VJ93 A0A0M3QZ91 E2ALM7 B3MXP7 B4R4B3 B4IGR0 X2JF59 Q9V3P0 A0A067QT52 G0WRL1 A0A411G805 A0A195DBS0 Q6XJ14 B3NWP9 A0A151I178 A0A158NMR0 A0A3B0JT65 L0ATY2 A0A151X141 A0A151IBS9 A0A0R3P480 Q29IR1 B4H2V8 A0A2J7QKP8 A0A232ET67 A0A0C9QZH8 T1E316 B4NBT6 F4WVS6 A0A1L8EF12 A0A0Q9X143 A0A151JTE8 B4L6U4 A0A023EJ61 K7J7Q6 A0A2P8Z9A3 A0A0J7L7W7 A0A0C9QCP4 A0A0K8TWN4 A0A034W382 A0A1L7LQE0 B1N694 U5EZ75 W8BQT7 A0A1I8Q0P3 Q17DN4 Q8WSF6 V5K6U7 A2I476 A0A0K8TUD5 A0A3P8ZAF2 A0A0A1WZP3 B4MA19 A0A182T5T8 A0A0F6MVY1 A0A182JGY2 A0A0B6Z6H1 A0A1L8EEU6 A0A0B6Z732 A0A3P9AQB1 A0A0B6Z4Q3 A0A3N0YJY2 I3PU24 T1DN15 A0A146J914 A0A2I8B5Y1 A0A1B6F8I1 A0A1W4WZJ8 A0A1B6INE9 B5XBY3 A0A1D2AJ30 W5LIA3 C1KC72 B5X7X7 A0A0M3SH29
B2KSE9 W0G2G4 A0A194QN77 A0A194Q0R0 I4DJ66 A0A3G1T1I6 A0A310SIG5 A0A0L7R4M0 A0A087ZND2 A0A154PDX5 E2BSB2 C3VVL4 A0A3L8DIR4 A0A026WMD6 A0A2A3E783 V9IJ36 T1P9R6 A0A0N0BCN5 A0A0L0C893 A0A1W4VJ93 A0A0M3QZ91 E2ALM7 B3MXP7 B4R4B3 B4IGR0 X2JF59 Q9V3P0 A0A067QT52 G0WRL1 A0A411G805 A0A195DBS0 Q6XJ14 B3NWP9 A0A151I178 A0A158NMR0 A0A3B0JT65 L0ATY2 A0A151X141 A0A151IBS9 A0A0R3P480 Q29IR1 B4H2V8 A0A2J7QKP8 A0A232ET67 A0A0C9QZH8 T1E316 B4NBT6 F4WVS6 A0A1L8EF12 A0A0Q9X143 A0A151JTE8 B4L6U4 A0A023EJ61 K7J7Q6 A0A2P8Z9A3 A0A0J7L7W7 A0A0C9QCP4 A0A0K8TWN4 A0A034W382 A0A1L7LQE0 B1N694 U5EZ75 W8BQT7 A0A1I8Q0P3 Q17DN4 Q8WSF6 V5K6U7 A2I476 A0A0K8TUD5 A0A3P8ZAF2 A0A0A1WZP3 B4MA19 A0A182T5T8 A0A0F6MVY1 A0A182JGY2 A0A0B6Z6H1 A0A1L8EEU6 A0A0B6Z732 A0A3P9AQB1 A0A0B6Z4Q3 A0A3N0YJY2 I3PU24 T1DN15 A0A146J914 A0A2I8B5Y1 A0A1B6F8I1 A0A1W4WZJ8 A0A1B6INE9 B5XBY3 A0A1D2AJ30 W5LIA3 C1KC72 B5X7X7 A0A0M3SH29
PDB
4XCS
E-value=1.80199e-84,
Score=792
Ontologies
GO
Topology
Subcellular location
Cytoplasm
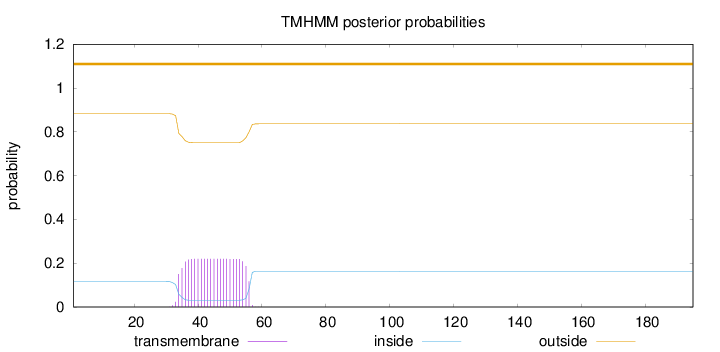
Length:
195
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
4.81683
Exp number, first 60 AAs:
4.81482
Total prob of N-in:
0.11698
outside
1 - 195
Population Genetic Test Statistics
Pi
348.779899
Theta
200.214408
Tajima's D
2.419239
CLR
0.464569
CSRT
0.936853157342133
Interpretation
Uncertain
