Gene
KWMTBOMO09351
Annotation
PREDICTED:_neuroligin-4?_Y-linked-like_[Bombyx_mori]
Full name
Carboxylic ester hydrolase
Location in the cell
Extracellular Reliability : 2.231
Sequence
CDS
ATGACCGTATTGGAAATAATAAAAGAGGGAACAAACTGTACTGTGAGTGTACGAGATGCTGTAGCCCGATATCCAGTGTTGGTCTTCGTCCACGGGGAGAGTTACGAGTGGAGTTCTGGAAACCCCTACGATGGGACAGTGTTGGCTTCTCACGCGGGACTAGTTGTGGTCACAATTAACTACAGACTTGGGATTTTGGGATTCCTCAACCCTAGATCCGATGACTACCCTCGGTCGCCAGCCAACTATGGTCTCATGGACCAAATTGCCGCCTTGCACTGGATCAAGGAAAACGTGGCGGTTTTTGGTGGAGATCCTACTAACGTCACTCTAATGGGACATGGCACCGGGGCTGCCTGCGTCCATTTCCTGCTCACCTCTTTAGCTGTGCCTGAAGCTACTGTCTGTTTTGCAGATGTCCAACACGAGATTTAA
Protein
MTVLEIIKEGTNCTVSVRDAVARYPVLVFVHGESYEWSSGNPYDGTVLASHAGLVVVTINYRLGILGFLNPRSDDYPRSPANYGLMDQIAALHWIKENVAVFGGDPTNVTLMGHGTGAACVHFLLTSLAVPEATVCFADVQHEI
Summary
Similarity
Belongs to the type-B carboxylesterase/lipase family.
Uniprot
A0A3S2NMG9
A0A2W1BB78
A0A2A4JUM4
A0A1L8D6I9
A0A194QTD3
A0A194Q1D9
+ More
E2AV44 N6U6A2 A0A1B6E2Z1 A0A0L7RFQ4 J9K9J8 U4U9V8 A0A1Y1NCU2 A0A067QT42 A0A151WUL2 A0A151JRS6 A0A151JVZ0 E2C2N5 A0A195AZ69 A0A195CX39 A0A139WHZ0 A0A026WI84 E0VI51 A0A2A3ES80 J9HF83 A0A0L0BN90 F4WI26 A0A2H8TED3 A0A087ZUT3 B9VMQ5 A0A1A9VW75 A4IJ79 K7J5U6 A0A1B6DJX8 A0A2S2NNP2 A0A2S2R616 B4N8A5 A0A068F5W8 Q29QD6 B3NYQ0 T1HEU8 W8CD45 B4I4V8 A0A023F4C2 B3LYX7 Q9VIC6 B4M4J0 A0A3B0JSU9 A0A182KAL3 B4K6S1 A0A1B6KRZ7 B4GMU8 A0A182H0Z7 A0A1W4V9Y5 B5DWB0 B4QZP7 A0A182LRF3 A0A0K8UBV6 B4JF78 B4PRG4 A0A1A9W1E9 W5JCT7 A0A1I8M0X6 A0A0M5IZD8 A0A1I8PVH1 A0A1S4G7C2 E9HH56 A0A2C9H8F4 A0A182VQ40 A0A182Y7E6 A0A182NLF9 A0A2P8Y3K3 A0A182JAK3 A0A0P5NQY5 A0A182Q9A1 A0A0N8BIS1 A0A1Y9HEJ0 A0A0P5ERE9 A0A0N8EFR4 A0A0P6GTR2 A0A182FAL5 A0A0P5IJN0 A0A0P6HEY2 A0A164RAU1 A0A0P5XSR9 A0A0P6BNN6 A0A0P5M6W2 A0A1Q3FUD5 A0A1S3DH91 A0A336MC47 A0A1B6KQL3 Q5TT55 E0VFS8 A0A0K2SYF8 A0A0J7K3X7 B0WIX9 A0A0A9ZGF2 A0A1J1I0C0 A0A1U9X1Y6
E2AV44 N6U6A2 A0A1B6E2Z1 A0A0L7RFQ4 J9K9J8 U4U9V8 A0A1Y1NCU2 A0A067QT42 A0A151WUL2 A0A151JRS6 A0A151JVZ0 E2C2N5 A0A195AZ69 A0A195CX39 A0A139WHZ0 A0A026WI84 E0VI51 A0A2A3ES80 J9HF83 A0A0L0BN90 F4WI26 A0A2H8TED3 A0A087ZUT3 B9VMQ5 A0A1A9VW75 A4IJ79 K7J5U6 A0A1B6DJX8 A0A2S2NNP2 A0A2S2R616 B4N8A5 A0A068F5W8 Q29QD6 B3NYQ0 T1HEU8 W8CD45 B4I4V8 A0A023F4C2 B3LYX7 Q9VIC6 B4M4J0 A0A3B0JSU9 A0A182KAL3 B4K6S1 A0A1B6KRZ7 B4GMU8 A0A182H0Z7 A0A1W4V9Y5 B5DWB0 B4QZP7 A0A182LRF3 A0A0K8UBV6 B4JF78 B4PRG4 A0A1A9W1E9 W5JCT7 A0A1I8M0X6 A0A0M5IZD8 A0A1I8PVH1 A0A1S4G7C2 E9HH56 A0A2C9H8F4 A0A182VQ40 A0A182Y7E6 A0A182NLF9 A0A2P8Y3K3 A0A182JAK3 A0A0P5NQY5 A0A182Q9A1 A0A0N8BIS1 A0A1Y9HEJ0 A0A0P5ERE9 A0A0N8EFR4 A0A0P6GTR2 A0A182FAL5 A0A0P5IJN0 A0A0P6HEY2 A0A164RAU1 A0A0P5XSR9 A0A0P6BNN6 A0A0P5M6W2 A0A1Q3FUD5 A0A1S3DH91 A0A336MC47 A0A1B6KQL3 Q5TT55 E0VFS8 A0A0K2SYF8 A0A0J7K3X7 B0WIX9 A0A0A9ZGF2 A0A1J1I0C0 A0A1U9X1Y6
EC Number
3.1.1.-
Pubmed
28756777
26354079
20798317
23537049
28004739
24845553
+ More
18362917 19820115 24508170 20566863 17510324 26108605 21719571 17069637 18974885 20075255 17994087 25880816 24495485 25474469 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26483478 15632085 17550304 20920257 23761445 25315136 21292972 25244985 29403074 12364791 14747013 17210077 25401762
18362917 19820115 24508170 20566863 17510324 26108605 21719571 17069637 18974885 20075255 17994087 25880816 24495485 25474469 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26483478 15632085 17550304 20920257 23761445 25315136 21292972 25244985 29403074 12364791 14747013 17210077 25401762
EMBL
RSAL01000253
RVE43466.1
KZ150175
PZC72572.1
NWSH01000626
PCG75183.1
+ More
GEYN01000068 JAV02061.1 KQ461195 KPJ06806.1 KQ459581 KPI99138.1 GL443020 EFN62695.1 APGK01037806 APGK01037807 APGK01037808 KB740949 ENN77160.1 GEDC01004998 JAS32300.1 KQ414602 KOC69648.1 ABLF02032237 ABLF02032238 ABLF02032239 ABLF02032241 KB631852 ERL86735.1 GEZM01006945 JAV95539.1 KK853310 KDR08664.1 KQ982727 KYQ51600.1 KQ978577 KYN30056.1 KQ981676 KYN38141.1 GL452201 EFN77791.1 KQ976697 KYM77538.1 KQ977231 KYN04724.1 KQ971342 KYB27546.1 KK107238 EZA54819.1 DS235184 EEB13057.1 KZ288193 PBC34132.1 CH477264 EJY57452.1 JRES01001621 KNC21491.1 GL888172 EGI66119.1 GFXV01000644 GFXV01005935 MBW12449.1 MBW17740.1 FJ580049 ACM48187.1 BT030437 ABO52857.1 AAZX01004420 AAZX01004645 AAZX01007714 AAZX01009171 AAZX01010230 AAZX01010833 AAZX01012199 AAZX01012817 AAZX01014915 AAZX01022750 GEDC01011305 JAS25993.1 GGMR01006184 MBY18803.1 GGMS01016248 MBY85451.1 CH964232 EDW81356.2 KJ702211 AID61365.1 BT024454 ABC86516.1 CH954181 EDV48163.1 ACPB03005614 GAMC01002586 JAC03970.1 CH480821 EDW55251.1 GBBI01002710 JAC16002.1 CH902617 EDV41851.2 KPU79385.1 KPU79386.1 AE014297 AAF53999.3 AAF54000.3 CH940652 EDW59551.2 OUUW01000008 SPP84093.1 CH933806 EDW14187.2 GEBQ01025751 JAT14226.1 CH479185 EDW38172.1 JXUM01102273 JXUM01102274 KQ564714 KXJ71855.1 CM000070 EDY68528.2 CM000364 EDX12005.1 AXCM01016645 GDHF01028241 JAI24073.1 CH916369 EDV93359.1 CM000160 EDW96352.2 ADMH02001760 ETN61158.1 CP012526 ALC45331.1 GL732645 EFX68935.1 PYGN01000971 PSN38825.1 GDIQ01143313 GDIQ01141464 JAL10262.1 AXCN02000135 GDIQ01173691 JAK78034.1 GDIP01156174 JAJ67228.1 GDIQ01031156 JAN63581.1 GDIQ01029609 JAN65128.1 GDIQ01214887 JAK36838.1 GDIQ01020416 JAN74321.1 LRGB01002190 KZS08483.1 GDIP01080073 JAM23642.1 GDIP01018760 JAM84955.1 GDIQ01159857 JAK91868.1 GFDL01003836 JAV31209.1 UFQS01000893 UFQT01000893 SSX07525.1 SSX27865.1 GEBQ01026256 JAT13721.1 AAAB01008888 EAL40590.4 DS235124 EEB12234.1 HACA01001453 CDW18814.1 LBMM01014885 KMQ85014.1 DS231953 EDS28805.1 GBHO01000458 JAG43146.1 CVRI01000024 CRK92270.1 KY021888 AQY62777.1
GEYN01000068 JAV02061.1 KQ461195 KPJ06806.1 KQ459581 KPI99138.1 GL443020 EFN62695.1 APGK01037806 APGK01037807 APGK01037808 KB740949 ENN77160.1 GEDC01004998 JAS32300.1 KQ414602 KOC69648.1 ABLF02032237 ABLF02032238 ABLF02032239 ABLF02032241 KB631852 ERL86735.1 GEZM01006945 JAV95539.1 KK853310 KDR08664.1 KQ982727 KYQ51600.1 KQ978577 KYN30056.1 KQ981676 KYN38141.1 GL452201 EFN77791.1 KQ976697 KYM77538.1 KQ977231 KYN04724.1 KQ971342 KYB27546.1 KK107238 EZA54819.1 DS235184 EEB13057.1 KZ288193 PBC34132.1 CH477264 EJY57452.1 JRES01001621 KNC21491.1 GL888172 EGI66119.1 GFXV01000644 GFXV01005935 MBW12449.1 MBW17740.1 FJ580049 ACM48187.1 BT030437 ABO52857.1 AAZX01004420 AAZX01004645 AAZX01007714 AAZX01009171 AAZX01010230 AAZX01010833 AAZX01012199 AAZX01012817 AAZX01014915 AAZX01022750 GEDC01011305 JAS25993.1 GGMR01006184 MBY18803.1 GGMS01016248 MBY85451.1 CH964232 EDW81356.2 KJ702211 AID61365.1 BT024454 ABC86516.1 CH954181 EDV48163.1 ACPB03005614 GAMC01002586 JAC03970.1 CH480821 EDW55251.1 GBBI01002710 JAC16002.1 CH902617 EDV41851.2 KPU79385.1 KPU79386.1 AE014297 AAF53999.3 AAF54000.3 CH940652 EDW59551.2 OUUW01000008 SPP84093.1 CH933806 EDW14187.2 GEBQ01025751 JAT14226.1 CH479185 EDW38172.1 JXUM01102273 JXUM01102274 KQ564714 KXJ71855.1 CM000070 EDY68528.2 CM000364 EDX12005.1 AXCM01016645 GDHF01028241 JAI24073.1 CH916369 EDV93359.1 CM000160 EDW96352.2 ADMH02001760 ETN61158.1 CP012526 ALC45331.1 GL732645 EFX68935.1 PYGN01000971 PSN38825.1 GDIQ01143313 GDIQ01141464 JAL10262.1 AXCN02000135 GDIQ01173691 JAK78034.1 GDIP01156174 JAJ67228.1 GDIQ01031156 JAN63581.1 GDIQ01029609 JAN65128.1 GDIQ01214887 JAK36838.1 GDIQ01020416 JAN74321.1 LRGB01002190 KZS08483.1 GDIP01080073 JAM23642.1 GDIP01018760 JAM84955.1 GDIQ01159857 JAK91868.1 GFDL01003836 JAV31209.1 UFQS01000893 UFQT01000893 SSX07525.1 SSX27865.1 GEBQ01026256 JAT13721.1 AAAB01008888 EAL40590.4 DS235124 EEB12234.1 HACA01001453 CDW18814.1 LBMM01014885 KMQ85014.1 DS231953 EDS28805.1 GBHO01000458 JAG43146.1 CVRI01000024 CRK92270.1 KY021888 AQY62777.1
Proteomes
UP000283053
UP000218220
UP000053240
UP000053268
UP000000311
UP000019118
+ More
UP000053825 UP000007819 UP000030742 UP000027135 UP000075809 UP000078492 UP000078541 UP000008237 UP000078540 UP000078542 UP000007266 UP000053097 UP000009046 UP000242457 UP000008820 UP000037069 UP000007755 UP000005203 UP000078200 UP000002358 UP000007798 UP000008711 UP000015103 UP000001292 UP000007801 UP000000803 UP000008792 UP000268350 UP000075881 UP000009192 UP000008744 UP000069940 UP000249989 UP000192221 UP000001819 UP000000304 UP000075883 UP000001070 UP000002282 UP000091820 UP000000673 UP000095301 UP000092553 UP000095300 UP000000305 UP000076407 UP000075920 UP000076408 UP000075884 UP000245037 UP000075880 UP000075886 UP000075900 UP000069272 UP000076858 UP000079169 UP000007062 UP000036403 UP000002320 UP000183832
UP000053825 UP000007819 UP000030742 UP000027135 UP000075809 UP000078492 UP000078541 UP000008237 UP000078540 UP000078542 UP000007266 UP000053097 UP000009046 UP000242457 UP000008820 UP000037069 UP000007755 UP000005203 UP000078200 UP000002358 UP000007798 UP000008711 UP000015103 UP000001292 UP000007801 UP000000803 UP000008792 UP000268350 UP000075881 UP000009192 UP000008744 UP000069940 UP000249989 UP000192221 UP000001819 UP000000304 UP000075883 UP000001070 UP000002282 UP000091820 UP000000673 UP000095301 UP000092553 UP000095300 UP000000305 UP000076407 UP000075920 UP000076408 UP000075884 UP000245037 UP000075880 UP000075886 UP000075900 UP000069272 UP000076858 UP000079169 UP000007062 UP000036403 UP000002320 UP000183832
Pfam
PF00135 COesterase
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
A0A3S2NMG9
A0A2W1BB78
A0A2A4JUM4
A0A1L8D6I9
A0A194QTD3
A0A194Q1D9
+ More
E2AV44 N6U6A2 A0A1B6E2Z1 A0A0L7RFQ4 J9K9J8 U4U9V8 A0A1Y1NCU2 A0A067QT42 A0A151WUL2 A0A151JRS6 A0A151JVZ0 E2C2N5 A0A195AZ69 A0A195CX39 A0A139WHZ0 A0A026WI84 E0VI51 A0A2A3ES80 J9HF83 A0A0L0BN90 F4WI26 A0A2H8TED3 A0A087ZUT3 B9VMQ5 A0A1A9VW75 A4IJ79 K7J5U6 A0A1B6DJX8 A0A2S2NNP2 A0A2S2R616 B4N8A5 A0A068F5W8 Q29QD6 B3NYQ0 T1HEU8 W8CD45 B4I4V8 A0A023F4C2 B3LYX7 Q9VIC6 B4M4J0 A0A3B0JSU9 A0A182KAL3 B4K6S1 A0A1B6KRZ7 B4GMU8 A0A182H0Z7 A0A1W4V9Y5 B5DWB0 B4QZP7 A0A182LRF3 A0A0K8UBV6 B4JF78 B4PRG4 A0A1A9W1E9 W5JCT7 A0A1I8M0X6 A0A0M5IZD8 A0A1I8PVH1 A0A1S4G7C2 E9HH56 A0A2C9H8F4 A0A182VQ40 A0A182Y7E6 A0A182NLF9 A0A2P8Y3K3 A0A182JAK3 A0A0P5NQY5 A0A182Q9A1 A0A0N8BIS1 A0A1Y9HEJ0 A0A0P5ERE9 A0A0N8EFR4 A0A0P6GTR2 A0A182FAL5 A0A0P5IJN0 A0A0P6HEY2 A0A164RAU1 A0A0P5XSR9 A0A0P6BNN6 A0A0P5M6W2 A0A1Q3FUD5 A0A1S3DH91 A0A336MC47 A0A1B6KQL3 Q5TT55 E0VFS8 A0A0K2SYF8 A0A0J7K3X7 B0WIX9 A0A0A9ZGF2 A0A1J1I0C0 A0A1U9X1Y6
E2AV44 N6U6A2 A0A1B6E2Z1 A0A0L7RFQ4 J9K9J8 U4U9V8 A0A1Y1NCU2 A0A067QT42 A0A151WUL2 A0A151JRS6 A0A151JVZ0 E2C2N5 A0A195AZ69 A0A195CX39 A0A139WHZ0 A0A026WI84 E0VI51 A0A2A3ES80 J9HF83 A0A0L0BN90 F4WI26 A0A2H8TED3 A0A087ZUT3 B9VMQ5 A0A1A9VW75 A4IJ79 K7J5U6 A0A1B6DJX8 A0A2S2NNP2 A0A2S2R616 B4N8A5 A0A068F5W8 Q29QD6 B3NYQ0 T1HEU8 W8CD45 B4I4V8 A0A023F4C2 B3LYX7 Q9VIC6 B4M4J0 A0A3B0JSU9 A0A182KAL3 B4K6S1 A0A1B6KRZ7 B4GMU8 A0A182H0Z7 A0A1W4V9Y5 B5DWB0 B4QZP7 A0A182LRF3 A0A0K8UBV6 B4JF78 B4PRG4 A0A1A9W1E9 W5JCT7 A0A1I8M0X6 A0A0M5IZD8 A0A1I8PVH1 A0A1S4G7C2 E9HH56 A0A2C9H8F4 A0A182VQ40 A0A182Y7E6 A0A182NLF9 A0A2P8Y3K3 A0A182JAK3 A0A0P5NQY5 A0A182Q9A1 A0A0N8BIS1 A0A1Y9HEJ0 A0A0P5ERE9 A0A0N8EFR4 A0A0P6GTR2 A0A182FAL5 A0A0P5IJN0 A0A0P6HEY2 A0A164RAU1 A0A0P5XSR9 A0A0P6BNN6 A0A0P5M6W2 A0A1Q3FUD5 A0A1S3DH91 A0A336MC47 A0A1B6KQL3 Q5TT55 E0VFS8 A0A0K2SYF8 A0A0J7K3X7 B0WIX9 A0A0A9ZGF2 A0A1J1I0C0 A0A1U9X1Y6
PDB
3BE8
E-value=3.17404e-28,
Score=305
Ontologies
GO
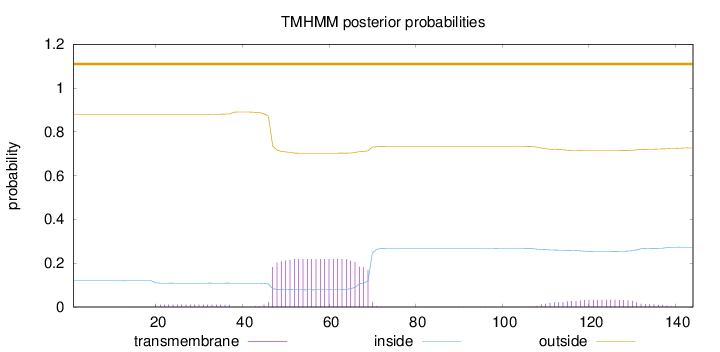
Topology
Length:
144
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.81873
Exp number, first 60 AAs:
3.23906
Total prob of N-in:
0.12007
outside
1 - 144
Population Genetic Test Statistics
Pi
238.306858
Theta
194.772686
Tajima's D
1.266896
CLR
0.356561
CSRT
0.731313434328284
Interpretation
Uncertain
