Gene
KWMTBOMO09348
Pre Gene Modal
BGIBMGA002170
Annotation
PREDICTED:_uncharacterized_protein_LOC101739297_[Bombyx_mori]
Full name
Carboxylic ester hydrolase
Location in the cell
Mitochondrial Reliability : 1.194
Sequence
CDS
ATGGCTTCTGACCGGCACGCGACTCCGTTTATAAAACAGAACGTGTTTATTTGTGTTGTTATCTCGCTCTTGCTCCCGAGGGTCAACGAAGGTGCGACAAAAATTTACGAGGGCAGCCGAGGTTTCCCGAACGATGACGGAGGGCGACGATTGACCCGAGAATACCATCTTCGGCAGGGAGCCTTGAGGGGACTGATAGTGAAACCAAATCGCCAGTATGATCTGCAGCTCGTGGAAATGTTTCTTGGCATACCTTATGCTGCCCCTCCTACTGGAAACTTCAGATTTATGCCGCCAGTCAGCGCTCCGCCCTGGCAAGGTGTTAGAATGGCCACTCATTTTGCCCCAGTGTGCCCTCAATCTCTTCCCCTAATAAAGAAGGGGAACCCGCCATCGCTCGGAAGACAGCGTTACCTCAACAAGCTGAAACCCTTTCTCTCGAATGAGAGTGAAGATTGCTTGTATCTTAACATCTACGTGCCGTATAGAGAACAAAAGTCCAAGAAGTTTCCCGTGTTGGTGTTTATACACGGTGACTCATTCGAGTGGAGCTCTGGGAATCCTTACGACGGAAGAATCTTATCCTCGTATGGGAACATCATGTTCGTAACAGTCAATTACCGACTCGGGATTTTAGGGTTCATGAAGCCAAGTCTGACGGAGCACGTGTACGGCAACAACGGTCTTCTGGATCAGTTGGCAGCGTTGCAGTGGATCAAGGATAACATTGAGGACTTGAACGGGGATCCGAACGCCGTGACCTTGATGGGGCACGGCACAGGAGCGGCTTCCGTCAACTATTTAATGCTCTCGCCGATATCAAACGGACTATTCCACCGGGCGATACTGATGTCAGGATCCGCCCTGTCAGACTGGGCTATGACGAGAGACCCGACTCAATACACCCTGCAGGTAGCCCAGAGCCTGGGTTGCAGCCCAAATTCGAAGAATTTGATGAACTGTATGCAGAAGAAGCCGCTATCAGAGATCAAGAAGGTGCAGATATTATCCAGAGAGTTTGAGACTCCGTTAGGTCCTGCAGTGGCCGGGTCGTTTGTACCGAGTGAGCCAGCGCAGACAATGGAAGCATTCCCAAATCTACTGAGCAAGTATCAATTACTGAGCGGAGTCACAGAATTGGAGAGCTATCACGATTTCGGGGTCATCGAACTGGATCACGGAATACTCGAGAATCAAAGGGACGATTTCATTAAAAAATACGCCAAGATCATCTTCGAAGGCGCCGAAAACGAAGCACTGAAAGAAATAATAAAAGAGTACGCGCCATCGAAACTAAACCCTGAAAGTTGGAGCGTGGAGAGCAACAGAGATTTAATACTGGATCTTCTCAGCGACGCGAGGACTTTATCGCCGGCGATACGCTTCGCCAACTACCACTCCGCCGCGAACCGGCAGTCTTACTTCTATGTGTTCGGACATAATTCTGTGAGCACGGAATATGCGCAGGTCAGTTTTGTCTAA
Protein
MASDRHATPFIKQNVFICVVISLLLPRVNEGATKIYEGSRGFPNDDGGRRLTREYHLRQGALRGLIVKPNRQYDLQLVEMFLGIPYAAPPTGNFRFMPPVSAPPWQGVRMATHFAPVCPQSLPLIKKGNPPSLGRQRYLNKLKPFLSNESEDCLYLNIYVPYREQKSKKFPVLVFIHGDSFEWSSGNPYDGRILSSYGNIMFVTVNYRLGILGFMKPSLTEHVYGNNGLLDQLAALQWIKDNIEDLNGDPNAVTLMGHGTGAASVNYLMLSPISNGLFHRAILMSGSALSDWAMTRDPTQYTLQVAQSLGCSPNSKNLMNCMQKKPLSEIKKVQILSREFETPLGPAVAGSFVPSEPAQTMEAFPNLLSKYQLLSGVTELESYHDFGVIELDHGILENQRDDFIKKYAKIIFEGAENEALKEIIKEYAPSKLNPESWSVESNRDLILDLLSDARTLSPAIRFANYHSAANRQSYFYVFGHNSVSTEYAQVSFV
Summary
Similarity
Belongs to the type-B carboxylesterase/lipase family.
Feature
chain Carboxylic ester hydrolase
Uniprot
A0A2W1BHR5
A0A0N1ICG6
A0A194Q0N0
A0A3S2L331
A0A0U2LN62
E0VI52
+ More
A0A182M3Z0 A0A1B0CTL9 A0A1W4VLM6 A0A1B6C6P7 A0A0R1E1E0 A0A182J161 A0A0R1E1R3 A0A084VBJ8 A0A1S4GID3 A0A182VQ42 A0A182Y7E4 A0A182K0R9 A0A182PS13 A0A182NLG0 A0A182QQP1 A0A182KXG8 A0A182VJD5 A0A182RZF6 A0A1I8JT78 A0A336LRV4 A0A182FAL3 W5JAX6 A0A0K8W6X0 A0A182H0H9 A0A0K8V4Y3 Q17GB8 A0A1S4F3S1 A0A1Q3G4H5 A0A1B0GQE1 A0A1Q3G493 A0A034W1A5 A0A1Q3G489 E9HH57 Q7QAR8 W8BHI7 A0A1W4VN12 Q9VIC7 B3NYQ3 B4I4W0 B3LYX9 A0A182WVU0 B4K6S2 A0A1J1I0C6 A0A0P6GKJ8 A0A0N8EFS0 A0A0P5M6G2 A0A0P6G820 A0A0P6DPP7 A0A0N8B9W5 A0A0P6FJW7 A0A0P5KXE6 A0A0P5Q7L5 A0A0N8EG89 A0A0P5QUF6 A0A0P5MA48 A0A0P6FFA3 A0A0P5ALC7 A0A0P5NL51 A0A0P6FQ59 A0A0N8BN44 A0A0P5GZ78 A0A0P6DXN6 A0A0P5RQX5 A0A0P6G3F7 A0A0P5JYT6 A0A0P6DGG2 A0A0P6FXX8 A0A0P5WPQ1 A0A0N8EA47 A0A0N8D3N3 A0A0P5X236 A0A154PNJ5 A0A0P5Y4J6 A0A0P5QHG2 A0A0P5I197 A0A0P5R442 A0A0P5RS49 B0WIY1 A0A0P5QHD0 B4N8A9 A0A0P5I5Q6 A0A0P5QM32 A0A0P5I874 A0A0P5WRZ1 A0A0N8DGS5 A0A0P5X2T9 A0A3B0JUL9 Q294N3 A0A1B6KQC4
A0A182M3Z0 A0A1B0CTL9 A0A1W4VLM6 A0A1B6C6P7 A0A0R1E1E0 A0A182J161 A0A0R1E1R3 A0A084VBJ8 A0A1S4GID3 A0A182VQ42 A0A182Y7E4 A0A182K0R9 A0A182PS13 A0A182NLG0 A0A182QQP1 A0A182KXG8 A0A182VJD5 A0A182RZF6 A0A1I8JT78 A0A336LRV4 A0A182FAL3 W5JAX6 A0A0K8W6X0 A0A182H0H9 A0A0K8V4Y3 Q17GB8 A0A1S4F3S1 A0A1Q3G4H5 A0A1B0GQE1 A0A1Q3G493 A0A034W1A5 A0A1Q3G489 E9HH57 Q7QAR8 W8BHI7 A0A1W4VN12 Q9VIC7 B3NYQ3 B4I4W0 B3LYX9 A0A182WVU0 B4K6S2 A0A1J1I0C6 A0A0P6GKJ8 A0A0N8EFS0 A0A0P5M6G2 A0A0P6G820 A0A0P6DPP7 A0A0N8B9W5 A0A0P6FJW7 A0A0P5KXE6 A0A0P5Q7L5 A0A0N8EG89 A0A0P5QUF6 A0A0P5MA48 A0A0P6FFA3 A0A0P5ALC7 A0A0P5NL51 A0A0P6FQ59 A0A0N8BN44 A0A0P5GZ78 A0A0P6DXN6 A0A0P5RQX5 A0A0P6G3F7 A0A0P5JYT6 A0A0P6DGG2 A0A0P6FXX8 A0A0P5WPQ1 A0A0N8EA47 A0A0N8D3N3 A0A0P5X236 A0A154PNJ5 A0A0P5Y4J6 A0A0P5QHG2 A0A0P5I197 A0A0P5R442 A0A0P5RS49 B0WIY1 A0A0P5QHD0 B4N8A9 A0A0P5I5Q6 A0A0P5QM32 A0A0P5I874 A0A0P5WRZ1 A0A0N8DGS5 A0A0P5X2T9 A0A3B0JUL9 Q294N3 A0A1B6KQC4
EC Number
3.1.1.-
Pubmed
EMBL
KZ150035
PZC74639.1
KQ459889
KPJ19593.1
KQ459581
KPI99137.1
+ More
RSAL01000197 RVE44542.1 KP308205 ALH43423.1 DS235184 EEB13058.1 AXCM01011415 AJWK01027772 AJWK01027773 AJWK01027774 AJWK01027775 GEDC01028151 JAS09147.1 CM000160 KRK03094.1 KRK03097.1 ATLV01007651 ATLV01007652 KE524424 KFB35342.1 AAAB01008888 AXCN02000135 APCN01005507 APCN01005508 UFQT01000083 SSX19453.1 ADMH02001760 ETN61156.1 GDHF01008479 GDHF01005443 JAI43835.1 JAI46871.1 JXUM01022712 JXUM01022713 JXUM01022714 JXUM01022715 JXUM01022716 JXUM01022717 JXUM01022718 KQ560639 KXJ81478.1 GDHF01025703 GDHF01018654 JAI26611.1 JAI33660.1 CH477264 EAT45587.1 GFDL01000351 JAV34694.1 AJVK01017029 AJVK01017030 AJVK01017031 AJVK01017032 AJVK01017033 GFDL01000427 JAV34618.1 GAKP01010840 JAC48112.1 GFDL01000428 JAV34617.1 GL732645 EFX68936.1 EAA08899.5 GAMC01013854 JAB92701.1 AE014297 BT024213 AAF53998.3 ABC86275.1 AFH06285.1 CH954181 EDV48166.1 CH480821 EDW55253.1 CH902617 EDV41853.1 KPU79389.1 KPU79390.1 CH933806 EDW14188.1 CVRI01000024 CRK92278.1 GDIQ01156103 GDIQ01091173 GDIQ01033154 GDIQ01032155 JAN62582.1 GDIQ01212782 GDIQ01153451 GDIQ01044991 GDIQ01031108 JAN63629.1 GDIQ01162849 JAK88876.1 GDIQ01048447 JAN46290.1 GDIQ01074479 JAN20258.1 GDIQ01229925 GDIQ01198540 GDIQ01198539 GDIQ01161509 GDIQ01161508 GDIQ01160001 GDIQ01160000 GDIQ01158205 GDIQ01137109 GDIQ01121561 JAK53186.1 GDIQ01048449 GDIQ01048448 JAN46288.1 GDIQ01177938 GDIQ01059985 JAK73787.1 GDIQ01139655 GDIQ01124015 JAL12071.1 GDIQ01029756 JAN64981.1 GDIQ01109513 JAL42213.1 GDIQ01162852 GDIQ01162851 GDIQ01162848 JAK88877.1 GDIQ01048446 JAN46291.1 GDIP01202308 JAJ21094.1 GDIQ01162850 JAK88875.1 GDIQ01061845 JAN32892.1 GDIQ01198538 GDIQ01161507 JAK90218.1 GDIQ01233774 GDIQ01233773 GDIQ01232023 GDIQ01127419 GDIQ01127418 GDIQ01125879 GDIQ01125878 JAK17951.1 GDIQ01071635 JAN23102.1 GDIQ01097275 GDIQ01097274 JAL54452.1 GDIQ01176290 GDIQ01173991 GDIQ01143158 GDIQ01143157 GDIQ01141462 GDIQ01141461 GDIQ01057538 JAN37199.1 GDIQ01197086 GDIQ01197085 JAK54639.1 GDIQ01079183 GDIQ01079182 JAN15555.1 GDIQ01042744 GDIQ01040096 JAN51993.1 GDIP01083331 JAM20384.1 GDIQ01046892 GDIQ01046891 JAN47845.1 GDIP01071983 LRGB01002190 JAM31732.1 KZS08485.1 GDIP01078525 JAM25190.1 KQ435007 KZC13419.1 GDIP01064181 JAM39534.1 GDIQ01217432 GDIQ01217429 GDIQ01114113 GDIQ01114110 JAL37613.1 GDIQ01219827 GDIQ01219825 JAK31900.1 GDIQ01115318 JAL36408.1 GDIQ01115317 JAL36409.1 DS231953 EDS28807.1 GDIQ01217431 GDIQ01214831 GDIQ01114112 JAL37614.1 CH964232 EDW81360.1 GDIQ01219826 JAK31899.1 GDIQ01115316 JAL36410.1 GDIQ01217430 GDIQ01114111 JAK34295.1 GDIP01083330 JAM20385.1 GDIP01035247 JAM68468.1 GDIP01078524 JAM25191.1 OUUW01000008 SPP84092.1 CM000070 EAL28931.3 GEBQ01026334 JAT13643.1
RSAL01000197 RVE44542.1 KP308205 ALH43423.1 DS235184 EEB13058.1 AXCM01011415 AJWK01027772 AJWK01027773 AJWK01027774 AJWK01027775 GEDC01028151 JAS09147.1 CM000160 KRK03094.1 KRK03097.1 ATLV01007651 ATLV01007652 KE524424 KFB35342.1 AAAB01008888 AXCN02000135 APCN01005507 APCN01005508 UFQT01000083 SSX19453.1 ADMH02001760 ETN61156.1 GDHF01008479 GDHF01005443 JAI43835.1 JAI46871.1 JXUM01022712 JXUM01022713 JXUM01022714 JXUM01022715 JXUM01022716 JXUM01022717 JXUM01022718 KQ560639 KXJ81478.1 GDHF01025703 GDHF01018654 JAI26611.1 JAI33660.1 CH477264 EAT45587.1 GFDL01000351 JAV34694.1 AJVK01017029 AJVK01017030 AJVK01017031 AJVK01017032 AJVK01017033 GFDL01000427 JAV34618.1 GAKP01010840 JAC48112.1 GFDL01000428 JAV34617.1 GL732645 EFX68936.1 EAA08899.5 GAMC01013854 JAB92701.1 AE014297 BT024213 AAF53998.3 ABC86275.1 AFH06285.1 CH954181 EDV48166.1 CH480821 EDW55253.1 CH902617 EDV41853.1 KPU79389.1 KPU79390.1 CH933806 EDW14188.1 CVRI01000024 CRK92278.1 GDIQ01156103 GDIQ01091173 GDIQ01033154 GDIQ01032155 JAN62582.1 GDIQ01212782 GDIQ01153451 GDIQ01044991 GDIQ01031108 JAN63629.1 GDIQ01162849 JAK88876.1 GDIQ01048447 JAN46290.1 GDIQ01074479 JAN20258.1 GDIQ01229925 GDIQ01198540 GDIQ01198539 GDIQ01161509 GDIQ01161508 GDIQ01160001 GDIQ01160000 GDIQ01158205 GDIQ01137109 GDIQ01121561 JAK53186.1 GDIQ01048449 GDIQ01048448 JAN46288.1 GDIQ01177938 GDIQ01059985 JAK73787.1 GDIQ01139655 GDIQ01124015 JAL12071.1 GDIQ01029756 JAN64981.1 GDIQ01109513 JAL42213.1 GDIQ01162852 GDIQ01162851 GDIQ01162848 JAK88877.1 GDIQ01048446 JAN46291.1 GDIP01202308 JAJ21094.1 GDIQ01162850 JAK88875.1 GDIQ01061845 JAN32892.1 GDIQ01198538 GDIQ01161507 JAK90218.1 GDIQ01233774 GDIQ01233773 GDIQ01232023 GDIQ01127419 GDIQ01127418 GDIQ01125879 GDIQ01125878 JAK17951.1 GDIQ01071635 JAN23102.1 GDIQ01097275 GDIQ01097274 JAL54452.1 GDIQ01176290 GDIQ01173991 GDIQ01143158 GDIQ01143157 GDIQ01141462 GDIQ01141461 GDIQ01057538 JAN37199.1 GDIQ01197086 GDIQ01197085 JAK54639.1 GDIQ01079183 GDIQ01079182 JAN15555.1 GDIQ01042744 GDIQ01040096 JAN51993.1 GDIP01083331 JAM20384.1 GDIQ01046892 GDIQ01046891 JAN47845.1 GDIP01071983 LRGB01002190 JAM31732.1 KZS08485.1 GDIP01078525 JAM25190.1 KQ435007 KZC13419.1 GDIP01064181 JAM39534.1 GDIQ01217432 GDIQ01217429 GDIQ01114113 GDIQ01114110 JAL37613.1 GDIQ01219827 GDIQ01219825 JAK31900.1 GDIQ01115318 JAL36408.1 GDIQ01115317 JAL36409.1 DS231953 EDS28807.1 GDIQ01217431 GDIQ01214831 GDIQ01114112 JAL37614.1 CH964232 EDW81360.1 GDIQ01219826 JAK31899.1 GDIQ01115316 JAL36410.1 GDIQ01217430 GDIQ01114111 JAK34295.1 GDIP01083330 JAM20385.1 GDIP01035247 JAM68468.1 GDIP01078524 JAM25191.1 OUUW01000008 SPP84092.1 CM000070 EAL28931.3 GEBQ01026334 JAT13643.1
Proteomes
UP000053240
UP000053268
UP000283053
UP000009046
UP000075883
UP000092461
+ More
UP000192221 UP000002282 UP000075880 UP000030765 UP000075920 UP000076408 UP000075881 UP000075885 UP000075884 UP000075886 UP000075882 UP000075903 UP000075900 UP000075840 UP000069272 UP000000673 UP000069940 UP000249989 UP000008820 UP000092462 UP000000305 UP000007062 UP000000803 UP000008711 UP000001292 UP000007801 UP000076407 UP000009192 UP000183832 UP000076858 UP000076502 UP000002320 UP000007798 UP000268350 UP000001819
UP000192221 UP000002282 UP000075880 UP000030765 UP000075920 UP000076408 UP000075881 UP000075885 UP000075884 UP000075886 UP000075882 UP000075903 UP000075900 UP000075840 UP000069272 UP000000673 UP000069940 UP000249989 UP000008820 UP000092462 UP000000305 UP000007062 UP000000803 UP000008711 UP000001292 UP000007801 UP000076407 UP000009192 UP000183832 UP000076858 UP000076502 UP000002320 UP000007798 UP000268350 UP000001819
PRIDE
Pfam
PF00135 COesterase
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
A0A2W1BHR5
A0A0N1ICG6
A0A194Q0N0
A0A3S2L331
A0A0U2LN62
E0VI52
+ More
A0A182M3Z0 A0A1B0CTL9 A0A1W4VLM6 A0A1B6C6P7 A0A0R1E1E0 A0A182J161 A0A0R1E1R3 A0A084VBJ8 A0A1S4GID3 A0A182VQ42 A0A182Y7E4 A0A182K0R9 A0A182PS13 A0A182NLG0 A0A182QQP1 A0A182KXG8 A0A182VJD5 A0A182RZF6 A0A1I8JT78 A0A336LRV4 A0A182FAL3 W5JAX6 A0A0K8W6X0 A0A182H0H9 A0A0K8V4Y3 Q17GB8 A0A1S4F3S1 A0A1Q3G4H5 A0A1B0GQE1 A0A1Q3G493 A0A034W1A5 A0A1Q3G489 E9HH57 Q7QAR8 W8BHI7 A0A1W4VN12 Q9VIC7 B3NYQ3 B4I4W0 B3LYX9 A0A182WVU0 B4K6S2 A0A1J1I0C6 A0A0P6GKJ8 A0A0N8EFS0 A0A0P5M6G2 A0A0P6G820 A0A0P6DPP7 A0A0N8B9W5 A0A0P6FJW7 A0A0P5KXE6 A0A0P5Q7L5 A0A0N8EG89 A0A0P5QUF6 A0A0P5MA48 A0A0P6FFA3 A0A0P5ALC7 A0A0P5NL51 A0A0P6FQ59 A0A0N8BN44 A0A0P5GZ78 A0A0P6DXN6 A0A0P5RQX5 A0A0P6G3F7 A0A0P5JYT6 A0A0P6DGG2 A0A0P6FXX8 A0A0P5WPQ1 A0A0N8EA47 A0A0N8D3N3 A0A0P5X236 A0A154PNJ5 A0A0P5Y4J6 A0A0P5QHG2 A0A0P5I197 A0A0P5R442 A0A0P5RS49 B0WIY1 A0A0P5QHD0 B4N8A9 A0A0P5I5Q6 A0A0P5QM32 A0A0P5I874 A0A0P5WRZ1 A0A0N8DGS5 A0A0P5X2T9 A0A3B0JUL9 Q294N3 A0A1B6KQC4
A0A182M3Z0 A0A1B0CTL9 A0A1W4VLM6 A0A1B6C6P7 A0A0R1E1E0 A0A182J161 A0A0R1E1R3 A0A084VBJ8 A0A1S4GID3 A0A182VQ42 A0A182Y7E4 A0A182K0R9 A0A182PS13 A0A182NLG0 A0A182QQP1 A0A182KXG8 A0A182VJD5 A0A182RZF6 A0A1I8JT78 A0A336LRV4 A0A182FAL3 W5JAX6 A0A0K8W6X0 A0A182H0H9 A0A0K8V4Y3 Q17GB8 A0A1S4F3S1 A0A1Q3G4H5 A0A1B0GQE1 A0A1Q3G493 A0A034W1A5 A0A1Q3G489 E9HH57 Q7QAR8 W8BHI7 A0A1W4VN12 Q9VIC7 B3NYQ3 B4I4W0 B3LYX9 A0A182WVU0 B4K6S2 A0A1J1I0C6 A0A0P6GKJ8 A0A0N8EFS0 A0A0P5M6G2 A0A0P6G820 A0A0P6DPP7 A0A0N8B9W5 A0A0P6FJW7 A0A0P5KXE6 A0A0P5Q7L5 A0A0N8EG89 A0A0P5QUF6 A0A0P5MA48 A0A0P6FFA3 A0A0P5ALC7 A0A0P5NL51 A0A0P6FQ59 A0A0N8BN44 A0A0P5GZ78 A0A0P6DXN6 A0A0P5RQX5 A0A0P6G3F7 A0A0P5JYT6 A0A0P6DGG2 A0A0P6FXX8 A0A0P5WPQ1 A0A0N8EA47 A0A0N8D3N3 A0A0P5X236 A0A154PNJ5 A0A0P5Y4J6 A0A0P5QHG2 A0A0P5I197 A0A0P5R442 A0A0P5RS49 B0WIY1 A0A0P5QHD0 B4N8A9 A0A0P5I5Q6 A0A0P5QM32 A0A0P5I874 A0A0P5WRZ1 A0A0N8DGS5 A0A0P5X2T9 A0A3B0JUL9 Q294N3 A0A1B6KQC4
PDB
5OJ6
E-value=8.6603e-74,
Score=705
Ontologies
GO
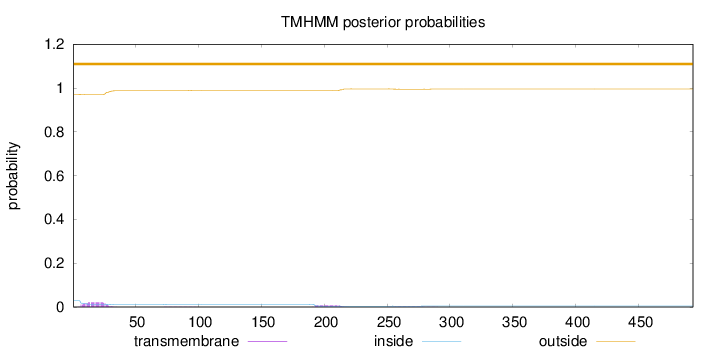
Topology
Length:
493
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.6478
Exp number, first 60 AAs:
0.41103
Total prob of N-in:
0.02850
outside
1 - 493
Population Genetic Test Statistics
Pi
265.223016
Theta
162.934772
Tajima's D
1.811047
CLR
0.719738
CSRT
0.851007449627519
Interpretation
Uncertain
