Gene
KWMTBOMO09345
Annotation
PREDICTED:_uncharacterized_protein_LOC106114667?_partial_[Papilio_xuthus]
Full name
Galactose-1-phosphate uridylyltransferase
Location in the cell
Nuclear Reliability : 3.591
Sequence
CDS
ATGGCGTCGCCTGTCGGGCCAGCGTGCCCGAATACAGAACTACCCTCAACACACATCAGGATACCACCGTTCTGGCCTGAAAAACCCGAAATATGGTTTGCGCAAGTGGAAGGACAATTTGCCATCATGCGAGTGACTGACGAGACGGCAAAATTTTATCATATATTGGCCAGCCTCGATCGCCAATATGCCGCCGAGGTAGAAGACGTGCTCACCGGTCCTCCCGATTACCAGCGGCTTAAAGAGGAGCTGATAAAAAGGTTGTCGGCGTCCCGCAGTAACAAGGTGAAGCAGCTATTGACGCACGAGCAGCTCGGTACCCGCAAACCGTCACAATTCTTGCGTCACCTACAACACCTCGCCGGACCGGAGATCCCGGAGGACTTCCTGCGGACTATATGGACCAGCCGGCTACCAACCACGTTGCAGCCGATCGTCGCGTCGCAGTCGACGCTTCCATTGGACGCACTCGCCGAGCTCGCCGACCGTGTCCACGAGATCGCAGCCCCAGTACACCACGTTGCGGCAGCCGTGACGTCATCATCATCATCGTCATCACAAATCGACAGGCTGTCCCAACAAGTAGCGGAGCTGACGCGGCAAGTCAGCGCCCTGACGACGCAGGTGCATCATCAACCGCGGCCGCAGGAGCGCGGGCGCGAACGTGGACGTCAACGACAACGCAGCCACTCATCCTCACGCCGGTCGCAGTCACACTACCGCAGGTACCCCATTTGCTGGTACCACAGCAAACACGGTGCCGAGGCGCGGAAGTGCGTGCGACCCTGCGACTACAAGGCGGGAAACCCGACAAGCAGGCAAGTGATGGCGACTCGCGACTGCCACAACTCTACGGGTCGCCTCTTCATCAAGGACCGCAGCAATAATATAGACTTCCTCATAGACACTGGAAGTGACATTTGTGTTTACCCTATTCATAAGTTGTGTGAACGCCGTGCTAAAACTAGTTATCAGCTCAGTGCTGCCAATGGTACAGTTATAGACACTTATGGTTATACGCAATTAAATGTTAATTTAGGCTTACGCCGCAACTTTCCATGGCGCTTTGTTGTTGCTGACGTCACAAAAGCCATTATAGGCGTCGATTTTTTGTCTCATTATCGACTCATAGTGGACGTCAGCCAACAAAGATTAATCGATAGTCTTACCAATATGAGCACTGTAGCCAGTCTAGATGTAAATAATAATCGTGTTACTAGTGTTAAGATAATGACAGGTGGCGACAGTAAGTACCACGCTATCTTGGCTGAATACCCGGAGCTAACACGCCCAGCTGGTACTCCAGGTATTGTCAAACACAACACGAAACACTACATACGCACTACACCAGGTCCACCCGTAGCATGCTCACCCCGTCGTCTCGCGCCGGACAAACTTAAAATTGCAAAACAAGAATTTAATATTATGATAGCCAACGGAACTGCACGTCCGTCTGACAGTCCATGGTCATCCCCGCTTCATCTTACTACCAAGAAGGACAACGGTTGGCGACCATGTGGTGATTACAGACAGTTAAATTCACGCACAATACCCGATCAGTATCCGATTCGTCATATACACGATTTTTCACAAAGTTTGTCTGGTTGTAAAATTTTTTCTGTCATCGATTTGGTCAAGGCCTATAATCAGATACCAACAGCGGAGGAGGATATCCCAAAGACCGCGATCACCACGCCATTTGCAAAACGCCACGTGACCACGAGGACCACTTGCGCCAACTTTTCAATCGTATGA
Protein
MASPVGPACPNTELPSTHIRIPPFWPEKPEIWFAQVEGQFAIMRVTDETAKFYHILASLDRQYAAEVEDVLTGPPDYQRLKEELIKRLSASRSNKVKQLLTHEQLGTRKPSQFLRHLQHLAGPEIPEDFLRTIWTSRLPTTLQPIVASQSTLPLDALAELADRVHEIAAPVHHVAAAVTSSSSSSSQIDRLSQQVAELTRQVSALTTQVHHQPRPQERGRERGRQRQRSHSSSRRSQSHYRRYPICWYHSKHGAEARKCVRPCDYKAGNPTSRQVMATRDCHNSTGRLFIKDRSNNIDFLIDTGSDICVYPIHKLCERRAKTSYQLSAANGTVIDTYGYTQLNVNLGLRRNFPWRFVVADVTKAIIGVDFLSHYRLIVDVSQQRLIDSLTNMSTVASLDVNNNRVTSVKIMTGGDSKYHAILAEYPELTRPAGTPGIVKHNTKHYIRTTPGPPVACSPRRLAPDKLKIAKQEFNIMIANGTARPSDSPWSSPLHLTTKKDNGWRPCGDYRQLNSRTIPDQYPIRHIHDFSQSLSGCKIFSVIDLVKAYNQIPTAEEDIPKTAITTPFAKRHVTTRTTCANFSIV
Summary
Catalytic Activity
alpha-D-galactose 1-phosphate + UDP-alpha-D-glucose = alpha-D-glucose 1-phosphate + UDP-alpha-D-galactose
Similarity
Belongs to the galactose-1-phosphate uridylyltransferase type 1 family.
Uniprot
A0A085NBV3
A0A139W8U7
D7GYF3
D7EJX5
C7C1Z9
D6WTN9
+ More
A0A139WA14 G7Y855 A0A0J7NWH7 G4VMU4 A0A085N387 Q9BM79 Q9BM81 W4ZGH9 A0A085LRV8 Q9BM80 G4VC96 A0A085MR14 A0A085N6R2 A0A0J7N0P4 A0A085N6P6 A0A085MSP0 X1XU27 A0A085ND37 X1XTW4 X1XU40 A0A3S2N6B3 A0A085MVG5 X1XPJ2 A0A0C2NDW5 X1X9N5 K7JAG6 A0A085LQK1 K7J8H5 X1XU13 A0A085LLJ9 J9LX06 A0A085MTV7 K7JAG8 K7JBH7 A0A3R7MAG3 A0A085M208 K7JCJ0 X1WVT2
A0A139WA14 G7Y855 A0A0J7NWH7 G4VMU4 A0A085N387 Q9BM79 Q9BM81 W4ZGH9 A0A085LRV8 Q9BM80 G4VC96 A0A085MR14 A0A085N6R2 A0A0J7N0P4 A0A085N6P6 A0A085MSP0 X1XU27 A0A085ND37 X1XTW4 X1XU40 A0A3S2N6B3 A0A085MVG5 X1XPJ2 A0A0C2NDW5 X1X9N5 K7JAG6 A0A085LQK1 K7J8H5 X1XU13 A0A085LLJ9 J9LX06 A0A085MTV7 K7JAG8 K7JBH7 A0A3R7MAG3 A0A085M208 K7JCJ0 X1WVT2
EC Number
2.7.7.12
EMBL
KL367519
KFD66949.1
KQ972957
KXZ75711.1
GG695519
EFA13383.1
+ More
KQ971759 EFA12902.2 FN356213 CAX83702.1 KQ971355 EFA07205.2 KQ971745 KYB24749.1 DF142933 GAA49140.1 LBMM01001181 KMQ96765.1 HE601630 CCD80981.1 KL367564 KFD63933.1 AY013569 AAK07487.1 AY013563 AAK07485.1 AAGJ04088930 KL363316 KFD47704.1 AY013565 AAK07486.1 HE601625 CCD77521.1 KL367778 KFD59660.1 KL367543 KFD65158.1 LBMM01012451 KMQ86215.1 KL367544 KFD65142.1 KL367683 KFD60236.1 ABLF02057446 ABLF02059301 KL367515 KFD67383.1 ABLF02005645 ABLF02042063 ABLF02040931 ABLF02040938 RSAL01000314 RVE42624.1 KL367631 KFD61211.1 ABLF02058233 JWZT01001363 KII72172.1 AAZX01023279 KL363334 KFD47247.1 AAZX01013940 ABLF02006413 ABLF02052270 KL363417 KFD45845.1 ABLF02031779 ABLF02031785 ABLF02043062 KL367655 KFD60653.1 AAZX01023264 QCYY01001531 ROT77388.1 KL363241 KFD51254.1 AAZX01006801 AAZX01023483 ABLF02003592 ABLF02059826 ABLF02066291
KQ971759 EFA12902.2 FN356213 CAX83702.1 KQ971355 EFA07205.2 KQ971745 KYB24749.1 DF142933 GAA49140.1 LBMM01001181 KMQ96765.1 HE601630 CCD80981.1 KL367564 KFD63933.1 AY013569 AAK07487.1 AY013563 AAK07485.1 AAGJ04088930 KL363316 KFD47704.1 AY013565 AAK07486.1 HE601625 CCD77521.1 KL367778 KFD59660.1 KL367543 KFD65158.1 LBMM01012451 KMQ86215.1 KL367544 KFD65142.1 KL367683 KFD60236.1 ABLF02057446 ABLF02059301 KL367515 KFD67383.1 ABLF02005645 ABLF02042063 ABLF02040931 ABLF02040938 RSAL01000314 RVE42624.1 KL367631 KFD61211.1 ABLF02058233 JWZT01001363 KII72172.1 AAZX01023279 KL363334 KFD47247.1 AAZX01013940 ABLF02006413 ABLF02052270 KL363417 KFD45845.1 ABLF02031779 ABLF02031785 ABLF02043062 KL367655 KFD60653.1 AAZX01023264 QCYY01001531 ROT77388.1 KL363241 KFD51254.1 AAZX01006801 AAZX01023483 ABLF02003592 ABLF02059826 ABLF02066291
Proteomes
Pfam
Interpro
IPR021109
Peptidase_aspartic_dom_sf
+ More
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR001995 Peptidase_A2_cat
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR000477 RT_dom
IPR001969 Aspartic_peptidase_AS
IPR041373 RT_RNaseH
IPR034132 RP_Saci-like
IPR041577 RT_RNaseH_2
IPR018289 MULE_transposase_dom
IPR005849 GalP_Utransf_N
IPR019779 GalP_UDPtransf1_His-AS
IPR001937 GalP_UDPtransf1
IPR036691 Endo/exonu/phosph_ase_sf
IPR036265 HIT-like_sf
IPR021896 Transposase_37
IPR041426 Mos1_HTH
IPR018061 Retropepsins
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR001995 Peptidase_A2_cat
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR000477 RT_dom
IPR001969 Aspartic_peptidase_AS
IPR041373 RT_RNaseH
IPR034132 RP_Saci-like
IPR041577 RT_RNaseH_2
IPR018289 MULE_transposase_dom
IPR005849 GalP_Utransf_N
IPR019779 GalP_UDPtransf1_His-AS
IPR001937 GalP_UDPtransf1
IPR036691 Endo/exonu/phosph_ase_sf
IPR036265 HIT-like_sf
IPR021896 Transposase_37
IPR041426 Mos1_HTH
IPR018061 Retropepsins
Gene 3D
ProteinModelPortal
A0A085NBV3
A0A139W8U7
D7GYF3
D7EJX5
C7C1Z9
D6WTN9
+ More
A0A139WA14 G7Y855 A0A0J7NWH7 G4VMU4 A0A085N387 Q9BM79 Q9BM81 W4ZGH9 A0A085LRV8 Q9BM80 G4VC96 A0A085MR14 A0A085N6R2 A0A0J7N0P4 A0A085N6P6 A0A085MSP0 X1XU27 A0A085ND37 X1XTW4 X1XU40 A0A3S2N6B3 A0A085MVG5 X1XPJ2 A0A0C2NDW5 X1X9N5 K7JAG6 A0A085LQK1 K7J8H5 X1XU13 A0A085LLJ9 J9LX06 A0A085MTV7 K7JAG8 K7JBH7 A0A3R7MAG3 A0A085M208 K7JCJ0 X1WVT2
A0A139WA14 G7Y855 A0A0J7NWH7 G4VMU4 A0A085N387 Q9BM79 Q9BM81 W4ZGH9 A0A085LRV8 Q9BM80 G4VC96 A0A085MR14 A0A085N6R2 A0A0J7N0P4 A0A085N6P6 A0A085MSP0 X1XU27 A0A085ND37 X1XTW4 X1XU40 A0A3S2N6B3 A0A085MVG5 X1XPJ2 A0A0C2NDW5 X1X9N5 K7JAG6 A0A085LQK1 K7J8H5 X1XU13 A0A085LLJ9 J9LX06 A0A085MTV7 K7JAG8 K7JBH7 A0A3R7MAG3 A0A085M208 K7JCJ0 X1WVT2
PDB
4OL8
E-value=5.36059e-14,
Score=190
Ontologies
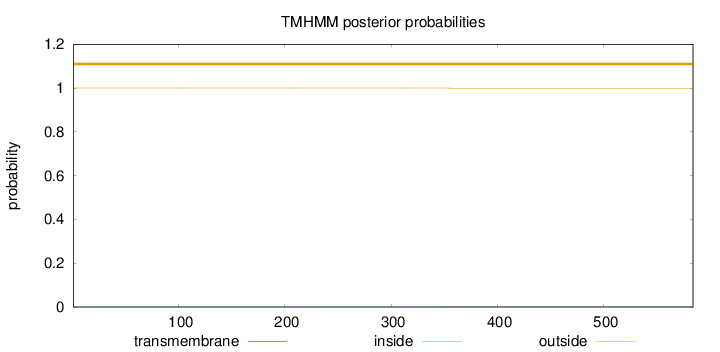
Topology
Length:
584
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01484
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00019
outside
1 - 584
Population Genetic Test Statistics
Pi
187.722025
Theta
26.64883
Tajima's D
4.206125
CLR
0
CSRT
0.999350032498375
Interpretation
Uncertain
