Gene
KWMTBOMO09342 Validated by peptides from experiments
Annotation
carboxypeptidase_inhibitor_precursor_[Bombyx_mori]
Full name
U-scoloptoxin(19)-Sm1a
+ More
U-scoloptoxin(19)-Tl1a
U-scoloptoxin(19)-Tl1a
Location in the cell
Extracellular Reliability : 4.016
Sequence
CDS
ATGAAGTTTTTATTATTTTGTGCAATGTGCATTTTAGTTTACGGAAACAGTGAAGATGATTTCTGTGAAATAGATTCCATTGAACAAGAGGATCCGTGCCGCAGGGAAGGTGGTTTGTGTACTGTGGCTGAGGACTGTCCATCTGATATCAGAGCCAGAACTGGACTCTGTCCTAAGCAGCAAAAAGACGGAATCGAATGCTGTTACGGAGTGTCCGTAAAAGAGACCAGATGTCGGAAGCACGGCGGAGAATGTTTCTCGAAAGGCTACTGCAGTCAATCTTTGATATATGAAGAAGCTTCAGACTGTCCCGAAGGGAATGATTGTTGTATTTTAGTGTAA
Protein
MKFLLFCAMCILVYGNSEDDFCEIDSIEQEDPCRREGGLCTVAEDCPSDIRARTGLCPKQQKDGIECCYGVSVKETRCRKHGGECFSKGYCSQSLIYEEASDCPEGNDCCILV
Summary
Similarity
Belongs to the scoloptoxin-19 family.
Keywords
Disulfide bond
Secreted
Signal
Toxin
Feature
chain U-scoloptoxin(19)-Sm1a
Uniprot
Q1HQ32
A0A2H1WLZ6
A0A437BB05
A0A2H1V3U0
A0A2W1BLL3
A0A194Q0Q0
+ More
A0A2A4J8Y0 A0A212EY05 A0A1B6DVH6 A0A2K8JVU2 A0A1I9WL85 I4DN50 A0A1Y1MH25 A0A1Y1MFS2 A0A0N1IQ18 A0A1L8DNU5 A0A1L8DNT2 A0A1L8D8P3 A0A1Y1MIS2 A0A1B0CKD4 A0A023EFP4 A0A023EFM7 Q16UL4 P0DQE9 A0A1Q3FQI1 A0A1Q3FQB9 A0A1Q3EUI8 A0A1Q3FQ82 A0A1Q3FQK5 A0A2W1BEQ4 B0WCP8 A0A1B0DK33 A0A226EUV8 T1IJ32 A0A182J734 A0A1I8PZN4 A0A0L0CML6 E0W0W7 A0A1B6KRN5 A0A023EGA1 Q9W2F6 A0A1Y1NLX5 T1PE04 B4J7L3 A0A0M4EXI1 A0A1W4W9K5 P0DQE8 B4MYP0 B4LLF2 A0A0U1TZE9 A0A0J9RJG4 Q16UL2 B3MC70 A0A182NJP4 Q28XM7 B4GH96 A0A0T6BFA1 A0A3B0JP46 A0A0R1DX67 T1E792 B3NMX9 A0A2M4AGN6 A0A2M4AGQ2 B4KRD3 A0A2M4DNU7 A0A2M3ZEF5 A0A2M3ZEG1 D3TSC2 A0A0N0PE82 A0A1B6IEP7 A0A182HWF4 A0NBL1 A0A0K8VVC2 A0A034VTU7 A0A084WJV5 A0A1A9UK29 W5JVU4 A0A0A1WH15 A0A182WWS4 A0A1B6EVB6 A0A182L398 A0A182RXP1 A0A182SG30 A0A182UQZ8 A0A087UQA0 A0A182JYH9 A0A182PF32 A0A2W1BIU1 A0A182TMN0 A0A182Y271 E9GD27 A0A212F444 A0A2M4C399
A0A2A4J8Y0 A0A212EY05 A0A1B6DVH6 A0A2K8JVU2 A0A1I9WL85 I4DN50 A0A1Y1MH25 A0A1Y1MFS2 A0A0N1IQ18 A0A1L8DNU5 A0A1L8DNT2 A0A1L8D8P3 A0A1Y1MIS2 A0A1B0CKD4 A0A023EFP4 A0A023EFM7 Q16UL4 P0DQE9 A0A1Q3FQI1 A0A1Q3FQB9 A0A1Q3EUI8 A0A1Q3FQ82 A0A1Q3FQK5 A0A2W1BEQ4 B0WCP8 A0A1B0DK33 A0A226EUV8 T1IJ32 A0A182J734 A0A1I8PZN4 A0A0L0CML6 E0W0W7 A0A1B6KRN5 A0A023EGA1 Q9W2F6 A0A1Y1NLX5 T1PE04 B4J7L3 A0A0M4EXI1 A0A1W4W9K5 P0DQE8 B4MYP0 B4LLF2 A0A0U1TZE9 A0A0J9RJG4 Q16UL2 B3MC70 A0A182NJP4 Q28XM7 B4GH96 A0A0T6BFA1 A0A3B0JP46 A0A0R1DX67 T1E792 B3NMX9 A0A2M4AGN6 A0A2M4AGQ2 B4KRD3 A0A2M4DNU7 A0A2M3ZEF5 A0A2M3ZEG1 D3TSC2 A0A0N0PE82 A0A1B6IEP7 A0A182HWF4 A0NBL1 A0A0K8VVC2 A0A034VTU7 A0A084WJV5 A0A1A9UK29 W5JVU4 A0A0A1WH15 A0A182WWS4 A0A1B6EVB6 A0A182L398 A0A182RXP1 A0A182SG30 A0A182UQZ8 A0A087UQA0 A0A182JYH9 A0A182PF32 A0A2W1BIU1 A0A182TMN0 A0A182Y271 E9GD27 A0A212F444 A0A2M4C399
Pubmed
28756777
26354079
22118469
27538518
22651552
28004739
+ More
24945155 17510324 24847043 26108605 20566863 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 17994087 22936249 15632085 17550304 20353571 12364791 14747013 17210077 25348373 24438588 20920257 23761445 25830018 20966253 25244985 21292972
24945155 17510324 24847043 26108605 20566863 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 17994087 22936249 15632085 17550304 20353571 12364791 14747013 17210077 25348373 24438588 20920257 23761445 25830018 20966253 25244985 21292972
EMBL
DQ443220
ABF51309.1
ODYU01009535
SOQ54037.1
RSAL01000099
RVE47590.1
+ More
ODYU01000555 SOQ35510.1 KZ150096 PZC73600.1 KQ459581 KPI99131.1 NWSH01002471 PCG68226.1 AGBW02011633 OWR46370.1 GEDC01007659 JAS29639.1 MF683289 ATU82430.1 KU932269 APA33905.1 AK402718 BAM19340.1 GEZM01035562 JAV83176.1 GEZM01035564 JAV83175.1 KQ460040 KPJ18377.1 GFDF01005952 JAV08132.1 GFDF01005951 JAV08133.1 GFDF01011238 JAV02846.1 GEZM01035561 JAV83177.1 AJWK01016026 GAPW01005958 GAPW01005955 GAPW01005953 JAC07640.1 GAPW01005957 JAC07641.1 CH477618 EAT38229.1 GFDL01005352 JAV29693.1 GFDL01005359 JAV29686.1 GFDL01016070 JAV18975.1 GFDL01005318 JAV29727.1 GFDL01005322 JAV29723.1 PZC73602.1 DS231889 EDS43699.1 AJVK01035228 AJVK01035229 LNIX01000001 OXA61372.1 JH430221 JRES01000182 KNC33545.1 DS235862 EEB19273.1 GEBQ01025897 JAT14080.1 GAPW01005956 GAPW01005954 JAC07644.1 AE013599 BT120316 AAF46736.1 ADC54207.1 GEZM01002423 JAV97266.1 KA646178 AFP60807.1 CH916367 EDW02161.1 CP012524 ALC42707.1 CH963894 EDW77229.1 CH940648 EDW61904.1 EU252478 ACD12022.1 CM002911 KMY95589.1 EAT38230.1 EAT38231.1 EAT38232.1 CH902619 EDV36170.1 CM000071 EAL26289.1 CH479183 EDW35866.1 LJIG01001381 KRT85559.1 OUUW01000001 SPP75359.1 CM000158 KRJ99715.1 GAMD01003252 JAA98338.1 CH954179 EDV55473.2 GGFK01006632 MBW39953.1 GGFK01006639 MBW39960.1 CH933808 EDW09349.1 GGFL01014610 MBW78788.1 GGFM01006128 MBW26879.1 GGFM01006124 MBW26875.1 EZ424324 EZ424325 ADD20600.1 ADD20601.1 KPJ18376.1 GECU01022290 JAS85416.1 APCN01001489 AAAB01008807 EAU77677.1 GDHF01019486 GDHF01009493 GDHF01003215 JAI32828.1 JAI42821.1 JAI49099.1 GAKP01013416 GAKP01013415 GAKP01013412 JAC45537.1 ATLV01024076 KE525348 KFB50499.1 ADMH02000263 ETN67195.1 GBXI01016306 JAC97985.1 GECZ01027900 JAS41869.1 KK121009 KFM79539.1 PZC73604.1 GL732539 EFX82765.1 AGBW02010437 OWR48518.1 GGFJ01010655 MBW59796.1
ODYU01000555 SOQ35510.1 KZ150096 PZC73600.1 KQ459581 KPI99131.1 NWSH01002471 PCG68226.1 AGBW02011633 OWR46370.1 GEDC01007659 JAS29639.1 MF683289 ATU82430.1 KU932269 APA33905.1 AK402718 BAM19340.1 GEZM01035562 JAV83176.1 GEZM01035564 JAV83175.1 KQ460040 KPJ18377.1 GFDF01005952 JAV08132.1 GFDF01005951 JAV08133.1 GFDF01011238 JAV02846.1 GEZM01035561 JAV83177.1 AJWK01016026 GAPW01005958 GAPW01005955 GAPW01005953 JAC07640.1 GAPW01005957 JAC07641.1 CH477618 EAT38229.1 GFDL01005352 JAV29693.1 GFDL01005359 JAV29686.1 GFDL01016070 JAV18975.1 GFDL01005318 JAV29727.1 GFDL01005322 JAV29723.1 PZC73602.1 DS231889 EDS43699.1 AJVK01035228 AJVK01035229 LNIX01000001 OXA61372.1 JH430221 JRES01000182 KNC33545.1 DS235862 EEB19273.1 GEBQ01025897 JAT14080.1 GAPW01005956 GAPW01005954 JAC07644.1 AE013599 BT120316 AAF46736.1 ADC54207.1 GEZM01002423 JAV97266.1 KA646178 AFP60807.1 CH916367 EDW02161.1 CP012524 ALC42707.1 CH963894 EDW77229.1 CH940648 EDW61904.1 EU252478 ACD12022.1 CM002911 KMY95589.1 EAT38230.1 EAT38231.1 EAT38232.1 CH902619 EDV36170.1 CM000071 EAL26289.1 CH479183 EDW35866.1 LJIG01001381 KRT85559.1 OUUW01000001 SPP75359.1 CM000158 KRJ99715.1 GAMD01003252 JAA98338.1 CH954179 EDV55473.2 GGFK01006632 MBW39953.1 GGFK01006639 MBW39960.1 CH933808 EDW09349.1 GGFL01014610 MBW78788.1 GGFM01006128 MBW26879.1 GGFM01006124 MBW26875.1 EZ424324 EZ424325 ADD20600.1 ADD20601.1 KPJ18376.1 GECU01022290 JAS85416.1 APCN01001489 AAAB01008807 EAU77677.1 GDHF01019486 GDHF01009493 GDHF01003215 JAI32828.1 JAI42821.1 JAI49099.1 GAKP01013416 GAKP01013415 GAKP01013412 JAC45537.1 ATLV01024076 KE525348 KFB50499.1 ADMH02000263 ETN67195.1 GBXI01016306 JAC97985.1 GECZ01027900 JAS41869.1 KK121009 KFM79539.1 PZC73604.1 GL732539 EFX82765.1 AGBW02010437 OWR48518.1 GGFJ01010655 MBW59796.1
Proteomes
UP000283053
UP000053268
UP000218220
UP000007151
UP000053240
UP000092461
+ More
UP000008820 UP000002320 UP000092462 UP000198287 UP000075880 UP000095300 UP000037069 UP000009046 UP000000803 UP000095301 UP000001070 UP000092553 UP000192221 UP000007798 UP000008792 UP000007801 UP000075884 UP000001819 UP000008744 UP000268350 UP000002282 UP000008711 UP000009192 UP000075840 UP000007062 UP000030765 UP000078200 UP000000673 UP000076407 UP000075882 UP000075900 UP000075901 UP000075903 UP000054359 UP000075881 UP000075885 UP000075902 UP000076408 UP000000305
UP000008820 UP000002320 UP000092462 UP000198287 UP000075880 UP000095300 UP000037069 UP000009046 UP000000803 UP000095301 UP000001070 UP000092553 UP000192221 UP000007798 UP000008792 UP000007801 UP000075884 UP000001819 UP000008744 UP000268350 UP000002282 UP000008711 UP000009192 UP000075840 UP000007062 UP000030765 UP000078200 UP000000673 UP000076407 UP000075882 UP000075900 UP000075901 UP000075903 UP000054359 UP000075881 UP000075885 UP000075902 UP000076408 UP000000305
Interpro
IPR023355
Myo_ane_neurotoxin_sf
Gene 3D
ProteinModelPortal
Q1HQ32
A0A2H1WLZ6
A0A437BB05
A0A2H1V3U0
A0A2W1BLL3
A0A194Q0Q0
+ More
A0A2A4J8Y0 A0A212EY05 A0A1B6DVH6 A0A2K8JVU2 A0A1I9WL85 I4DN50 A0A1Y1MH25 A0A1Y1MFS2 A0A0N1IQ18 A0A1L8DNU5 A0A1L8DNT2 A0A1L8D8P3 A0A1Y1MIS2 A0A1B0CKD4 A0A023EFP4 A0A023EFM7 Q16UL4 P0DQE9 A0A1Q3FQI1 A0A1Q3FQB9 A0A1Q3EUI8 A0A1Q3FQ82 A0A1Q3FQK5 A0A2W1BEQ4 B0WCP8 A0A1B0DK33 A0A226EUV8 T1IJ32 A0A182J734 A0A1I8PZN4 A0A0L0CML6 E0W0W7 A0A1B6KRN5 A0A023EGA1 Q9W2F6 A0A1Y1NLX5 T1PE04 B4J7L3 A0A0M4EXI1 A0A1W4W9K5 P0DQE8 B4MYP0 B4LLF2 A0A0U1TZE9 A0A0J9RJG4 Q16UL2 B3MC70 A0A182NJP4 Q28XM7 B4GH96 A0A0T6BFA1 A0A3B0JP46 A0A0R1DX67 T1E792 B3NMX9 A0A2M4AGN6 A0A2M4AGQ2 B4KRD3 A0A2M4DNU7 A0A2M3ZEF5 A0A2M3ZEG1 D3TSC2 A0A0N0PE82 A0A1B6IEP7 A0A182HWF4 A0NBL1 A0A0K8VVC2 A0A034VTU7 A0A084WJV5 A0A1A9UK29 W5JVU4 A0A0A1WH15 A0A182WWS4 A0A1B6EVB6 A0A182L398 A0A182RXP1 A0A182SG30 A0A182UQZ8 A0A087UQA0 A0A182JYH9 A0A182PF32 A0A2W1BIU1 A0A182TMN0 A0A182Y271 E9GD27 A0A212F444 A0A2M4C399
A0A2A4J8Y0 A0A212EY05 A0A1B6DVH6 A0A2K8JVU2 A0A1I9WL85 I4DN50 A0A1Y1MH25 A0A1Y1MFS2 A0A0N1IQ18 A0A1L8DNU5 A0A1L8DNT2 A0A1L8D8P3 A0A1Y1MIS2 A0A1B0CKD4 A0A023EFP4 A0A023EFM7 Q16UL4 P0DQE9 A0A1Q3FQI1 A0A1Q3FQB9 A0A1Q3EUI8 A0A1Q3FQ82 A0A1Q3FQK5 A0A2W1BEQ4 B0WCP8 A0A1B0DK33 A0A226EUV8 T1IJ32 A0A182J734 A0A1I8PZN4 A0A0L0CML6 E0W0W7 A0A1B6KRN5 A0A023EGA1 Q9W2F6 A0A1Y1NLX5 T1PE04 B4J7L3 A0A0M4EXI1 A0A1W4W9K5 P0DQE8 B4MYP0 B4LLF2 A0A0U1TZE9 A0A0J9RJG4 Q16UL2 B3MC70 A0A182NJP4 Q28XM7 B4GH96 A0A0T6BFA1 A0A3B0JP46 A0A0R1DX67 T1E792 B3NMX9 A0A2M4AGN6 A0A2M4AGQ2 B4KRD3 A0A2M4DNU7 A0A2M3ZEF5 A0A2M3ZEG1 D3TSC2 A0A0N0PE82 A0A1B6IEP7 A0A182HWF4 A0NBL1 A0A0K8VVC2 A0A034VTU7 A0A084WJV5 A0A1A9UK29 W5JVU4 A0A0A1WH15 A0A182WWS4 A0A1B6EVB6 A0A182L398 A0A182RXP1 A0A182SG30 A0A182UQZ8 A0A087UQA0 A0A182JYH9 A0A182PF32 A0A2W1BIU1 A0A182TMN0 A0A182Y271 E9GD27 A0A212F444 A0A2M4C399
Ontologies
Topology
Subcellular location
Secreted
SignalP
Position: 1 - 17,
Likelihood: 0.999547
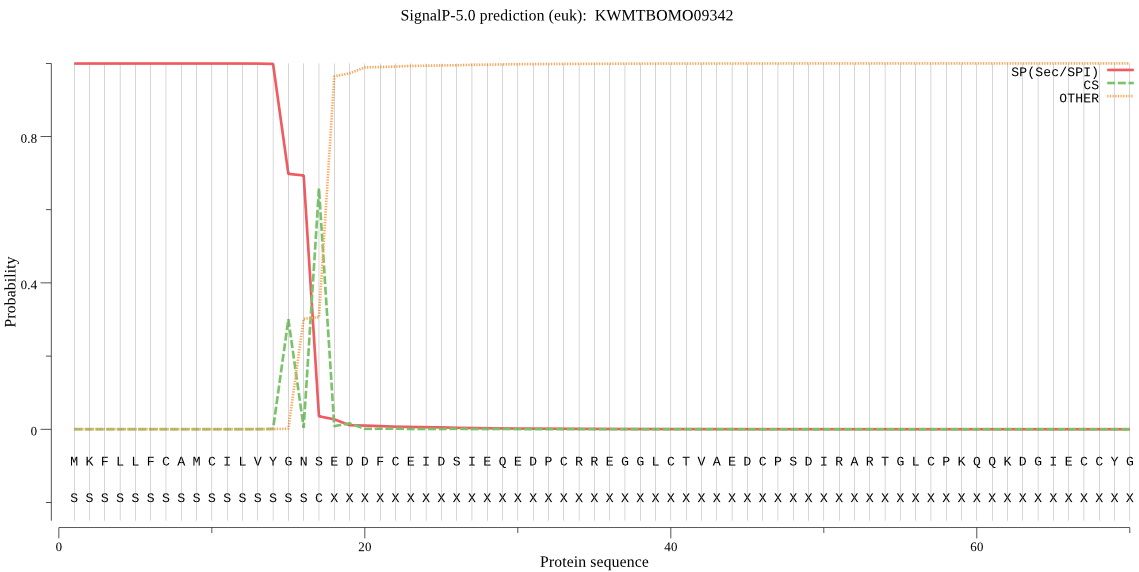
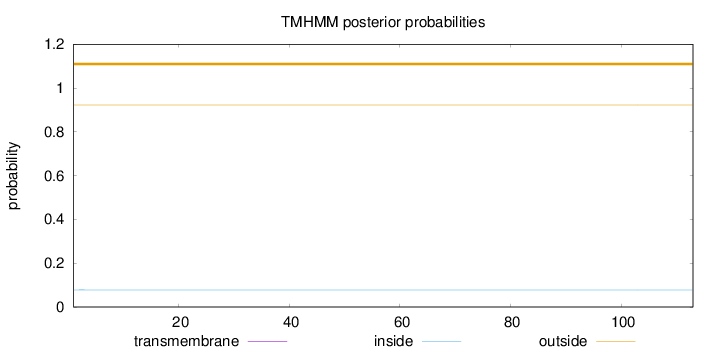
Length:
113
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00046
Exp number, first 60 AAs:
0.00046
Total prob of N-in:
0.07800
outside
1 - 113
Population Genetic Test Statistics
Pi
111.899583
Theta
10.607099
Tajima's D
-1.138226
CLR
0.233737
CSRT
0.111794410279486
Interpretation
Uncertain
