Gene
KWMTBOMO09337
Pre Gene Modal
BGIBMGA002172
Annotation
PREDICTED:_neuroligin-4?_X-linked-like_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.59
Sequence
CDS
ATGCTGAGGACCAGAGTAATCGGCACTCGCTACGGAAAACTCCAAGGGGTAATTTTACCTATGGATCAGCACAAGTACTTGAAACCTGTCGAGGCGTACCTCGGGGTGCCATATGCCACACCACCCACGGGTTCTAACAGATTTGCGCCAACACGAGCTCCAGCACCATGGGATGAGGTGAAAATGGTGGACCAAATGGGACCAGTCTGCCCACAGAGACTGCCGGATATTTCGAACGAGACCTTAGCTTTAGAGAGAATGCCTAAGGGACGTCTGGAATACTTAAGGAGATTGCTTCCTCGGCTTAAGAATCAGAGTGAAGACTGTCTATATATGAACATCTATACACCAGTTCAAGTTGGGCCAACGCTCCAAGCCAAGTACCCAGTAATCGTTTTCATTCACGGGGAGTCTTTCGAATGGAATTCCGGTAATGTTTACGATGGCTCCGTGTTGGCTAGCTACGCTGGATTAGTTGTTATCACCATCAATTACAGGCTCGGTATCCTTGGTAAGTATAATAATACTTTTGGATAA
Protein
MLRTRVIGTRYGKLQGVILPMDQHKYLKPVEAYLGVPYATPPTGSNRFAPTRAPAPWDEVKMVDQMGPVCPQRLPDISNETLALERMPKGRLEYLRRLLPRLKNQSEDCLYMNIYTPVQVGPTLQAKYPVIVFIHGESFEWNSGNVYDGSVLASYAGLVVITINYRLGILGKYNNTFG
Summary
Uniprot
A0A437BB32
A0A2W1BDC8
A0A194RMK8
A0A194Q2A5
A0A212EHX5
A0A1L8D6N7
+ More
A0A139WI26 A0A1I8PGZ6 E0VFS8 A0A1I8MUT4 A0A1B0AJ85 A0A1A9X8I2 A0A1B0B268 Q7PGX1 A0A1A9X427 B4GL70 A0A068F5U0 B4QS57 B3NZ06 A0A0L0BSZ4 B4IUR7 A0A0P8XXC1 B4LXI9 A0A0A9ZGF2 B4IIE9 Q9VDP5 B4K9C7 B6IDZ4 A0A2H1VKL6 A0A0Q9X167 A0A1W4VXD5 W8BSI7 A0A0Q9X8Z0 B5DVH8 A0A3B0KJP9 B4NGA7 A0A232FEM3 A0A0M5J2B0 B4JTH3 H9IY47 A0A437BB17 A0A151JW79 A0A2S2PN03 A0A2S2QDT7 A0A087ZVK3 B9VMQ6 E0VI51 A0A2H1V8H5 A0A0U2LHT4 A0A2A4K584 A0A2W1BEL2 A0A1B0CSA5 A0A1B6CJ17 A0A1B6MDA4 B4M4J0 A0A2C9H8F4 A0A1S4G7C2 A0A1Y9HEJ0 A0A212FJJ8 A0A023F4C2 A0A182Q9A1 A0A1Q3FUD5 A0A194QTD3 A0A2A3E952 A0A2W1BB78 A0A1L8D6I9 A0A087ZUT3 B9VMQ5 A0A2A4JUM4 A0A0M5IZD8 T1L1F5 A0A194Q1D9 A0A1B6KRZ7 A0A2S2R616 E9HH57 E9HH56 Q5TT55 A0A0N8EFR4 A0A1B6D837 A0A164RAU1 A0A0P5XSR9 A0A0P5IJN0 A0A0P6GTR2 A0A0P5MA48 A0A0N8EA47 A0A0P6FXX8 A0A0P5M6G2 A0A0P6FFA3 A0A0P5NL51 A0A0P5QUF6 A0A0P6FQ59 A0A0P6G3F7 A0A0P5RQX5 A0A0P6DGG2 A0A0P6G820 A0A0P6FJW7 A0A0P5WPQ1
A0A139WI26 A0A1I8PGZ6 E0VFS8 A0A1I8MUT4 A0A1B0AJ85 A0A1A9X8I2 A0A1B0B268 Q7PGX1 A0A1A9X427 B4GL70 A0A068F5U0 B4QS57 B3NZ06 A0A0L0BSZ4 B4IUR7 A0A0P8XXC1 B4LXI9 A0A0A9ZGF2 B4IIE9 Q9VDP5 B4K9C7 B6IDZ4 A0A2H1VKL6 A0A0Q9X167 A0A1W4VXD5 W8BSI7 A0A0Q9X8Z0 B5DVH8 A0A3B0KJP9 B4NGA7 A0A232FEM3 A0A0M5J2B0 B4JTH3 H9IY47 A0A437BB17 A0A151JW79 A0A2S2PN03 A0A2S2QDT7 A0A087ZVK3 B9VMQ6 E0VI51 A0A2H1V8H5 A0A0U2LHT4 A0A2A4K584 A0A2W1BEL2 A0A1B0CSA5 A0A1B6CJ17 A0A1B6MDA4 B4M4J0 A0A2C9H8F4 A0A1S4G7C2 A0A1Y9HEJ0 A0A212FJJ8 A0A023F4C2 A0A182Q9A1 A0A1Q3FUD5 A0A194QTD3 A0A2A3E952 A0A2W1BB78 A0A1L8D6I9 A0A087ZUT3 B9VMQ5 A0A2A4JUM4 A0A0M5IZD8 T1L1F5 A0A194Q1D9 A0A1B6KRZ7 A0A2S2R616 E9HH57 E9HH56 Q5TT55 A0A0N8EFR4 A0A1B6D837 A0A164RAU1 A0A0P5XSR9 A0A0P5IJN0 A0A0P6GTR2 A0A0P5MA48 A0A0N8EA47 A0A0P6FXX8 A0A0P5M6G2 A0A0P6FFA3 A0A0P5NL51 A0A0P5QUF6 A0A0P6FQ59 A0A0P6G3F7 A0A0P5RQX5 A0A0P6DGG2 A0A0P6G820 A0A0P6FJW7 A0A0P5WPQ1
Pubmed
28756777
26354079
22118469
18362917
19820115
20566863
+ More
25315136 12364791 14747013 17210077 17994087 25880816 26108605 17550304 25401762 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 24495485 15632085 28648823 19121390 17069637 18974885 26749290 17510324 25474469 21292972
25315136 12364791 14747013 17210077 17994087 25880816 26108605 17550304 25401762 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 24495485 15632085 28648823 19121390 17069637 18974885 26749290 17510324 25474469 21292972
EMBL
RSAL01000099
RVE47591.1
KZ150139
PZC72958.1
KQ460154
KPJ17231.1
+ More
KQ459581 KPI99129.1 AGBW02014763 OWR41088.1 GEYN01000074 JAV02055.1 KQ971342 KYB27544.1 DS235124 EEB12234.1 JXJN01007432 AAAB01008879 EAA44773.5 CH479185 EDW38294.1 KJ702186 AID61340.1 CM000364 EDX12253.1 CH954181 EDV48409.1 JRES01001389 KNC23200.1 CH892041 EDX00131.2 CH902617 KPU79345.1 CH940650 EDW66773.2 GBHO01000458 JAG43146.1 CH480842 EDW49693.1 AE014297 AAF55745.4 ACZ94957.1 AHN57419.1 CH933806 EDW15559.2 BT050584 ACJ23468.1 ACZ94958.1 ODYU01003066 SOQ41368.1 KRG01621.1 KRG01622.1 GAMC01006682 GAMC01006681 JAB99873.1 KRG01620.1 CM000070 EDY68564.3 OUUW01000008 SPP83978.1 CH964251 EDW83324.2 NNAY01000326 OXU29206.1 CP012526 ALC46886.1 CH916373 EDV95063.1 BABH01029721 BABH01029722 BABH01029723 BABH01029724 RVE47584.1 KQ981676 KYN38135.1 GGMR01018148 MBY30767.1 GGMS01006704 MBY75907.1 FJ580050 ACM48188.1 DS235184 EEB13057.1 ODYU01001227 SOQ37150.1 KP308206 ALH43424.1 NWSH01000115 PCG79397.1 KZ150096 PZC73599.1 AJWK01025848 AJWK01025849 AJWK01025850 AJWK01025851 AJWK01025852 AJWK01025853 AJWK01025854 AJWK01025855 GEDC01023812 JAS13486.1 GEBQ01006118 JAT33859.1 CH940652 EDW59551.2 AGBW02008258 OWR53904.1 GBBI01002710 JAC16002.1 AXCN02000135 GFDL01003836 JAV31209.1 KQ461195 KPJ06806.1 KZ288345 PBC27591.1 KZ150175 PZC72572.1 GEYN01000068 JAV02061.1 FJ580049 ACM48187.1 NWSH01000626 PCG75183.1 ALC45331.1 CAEY01000897 CAEY01000898 KPI99138.1 GEBQ01025751 JAT14226.1 GGMS01016248 MBY85451.1 GL732645 EFX68936.1 EFX68935.1 AAAB01008888 EAL40590.4 GDIQ01031156 JAN63581.1 GEDC01015441 JAS21857.1 LRGB01002190 KZS08483.1 GDIP01080073 JAM23642.1 GDIQ01214887 JAK36838.1 GDIQ01029609 JAN65128.1 GDIQ01162852 GDIQ01162851 GDIQ01162848 JAK88877.1 GDIQ01046892 GDIQ01046891 JAN47845.1 GDIQ01042744 GDIQ01040096 JAN51993.1 GDIQ01162849 JAK88876.1 GDIQ01048446 JAN46291.1 GDIQ01162850 JAK88875.1 GDIQ01109513 JAL42213.1 GDIQ01061845 JAN32892.1 GDIQ01176290 GDIQ01173991 GDIQ01143158 GDIQ01143157 GDIQ01141462 GDIQ01141461 GDIQ01057538 JAN37199.1 GDIQ01097275 GDIQ01097274 JAL54452.1 GDIQ01079183 GDIQ01079182 JAN15555.1 GDIQ01048447 JAN46290.1 GDIQ01048449 GDIQ01048448 JAN46288.1 GDIP01083331 JAM20384.1
KQ459581 KPI99129.1 AGBW02014763 OWR41088.1 GEYN01000074 JAV02055.1 KQ971342 KYB27544.1 DS235124 EEB12234.1 JXJN01007432 AAAB01008879 EAA44773.5 CH479185 EDW38294.1 KJ702186 AID61340.1 CM000364 EDX12253.1 CH954181 EDV48409.1 JRES01001389 KNC23200.1 CH892041 EDX00131.2 CH902617 KPU79345.1 CH940650 EDW66773.2 GBHO01000458 JAG43146.1 CH480842 EDW49693.1 AE014297 AAF55745.4 ACZ94957.1 AHN57419.1 CH933806 EDW15559.2 BT050584 ACJ23468.1 ACZ94958.1 ODYU01003066 SOQ41368.1 KRG01621.1 KRG01622.1 GAMC01006682 GAMC01006681 JAB99873.1 KRG01620.1 CM000070 EDY68564.3 OUUW01000008 SPP83978.1 CH964251 EDW83324.2 NNAY01000326 OXU29206.1 CP012526 ALC46886.1 CH916373 EDV95063.1 BABH01029721 BABH01029722 BABH01029723 BABH01029724 RVE47584.1 KQ981676 KYN38135.1 GGMR01018148 MBY30767.1 GGMS01006704 MBY75907.1 FJ580050 ACM48188.1 DS235184 EEB13057.1 ODYU01001227 SOQ37150.1 KP308206 ALH43424.1 NWSH01000115 PCG79397.1 KZ150096 PZC73599.1 AJWK01025848 AJWK01025849 AJWK01025850 AJWK01025851 AJWK01025852 AJWK01025853 AJWK01025854 AJWK01025855 GEDC01023812 JAS13486.1 GEBQ01006118 JAT33859.1 CH940652 EDW59551.2 AGBW02008258 OWR53904.1 GBBI01002710 JAC16002.1 AXCN02000135 GFDL01003836 JAV31209.1 KQ461195 KPJ06806.1 KZ288345 PBC27591.1 KZ150175 PZC72572.1 GEYN01000068 JAV02061.1 FJ580049 ACM48187.1 NWSH01000626 PCG75183.1 ALC45331.1 CAEY01000897 CAEY01000898 KPI99138.1 GEBQ01025751 JAT14226.1 GGMS01016248 MBY85451.1 GL732645 EFX68936.1 EFX68935.1 AAAB01008888 EAL40590.4 GDIQ01031156 JAN63581.1 GEDC01015441 JAS21857.1 LRGB01002190 KZS08483.1 GDIP01080073 JAM23642.1 GDIQ01214887 JAK36838.1 GDIQ01029609 JAN65128.1 GDIQ01162852 GDIQ01162851 GDIQ01162848 JAK88877.1 GDIQ01046892 GDIQ01046891 JAN47845.1 GDIQ01042744 GDIQ01040096 JAN51993.1 GDIQ01162849 JAK88876.1 GDIQ01048446 JAN46291.1 GDIQ01162850 JAK88875.1 GDIQ01109513 JAL42213.1 GDIQ01061845 JAN32892.1 GDIQ01176290 GDIQ01173991 GDIQ01143158 GDIQ01143157 GDIQ01141462 GDIQ01141461 GDIQ01057538 JAN37199.1 GDIQ01097275 GDIQ01097274 JAL54452.1 GDIQ01079183 GDIQ01079182 JAN15555.1 GDIQ01048447 JAN46290.1 GDIQ01048449 GDIQ01048448 JAN46288.1 GDIP01083331 JAM20384.1
Proteomes
UP000283053
UP000053240
UP000053268
UP000007151
UP000007266
UP000095300
+ More
UP000009046 UP000095301 UP000092445 UP000092443 UP000092460 UP000007062 UP000091820 UP000008744 UP000000304 UP000008711 UP000037069 UP000002282 UP000007801 UP000008792 UP000001292 UP000000803 UP000009192 UP000192221 UP000001819 UP000268350 UP000007798 UP000215335 UP000092553 UP000001070 UP000005204 UP000078541 UP000005203 UP000218220 UP000092461 UP000076407 UP000075900 UP000075886 UP000242457 UP000015104 UP000000305 UP000076858
UP000009046 UP000095301 UP000092445 UP000092443 UP000092460 UP000007062 UP000091820 UP000008744 UP000000304 UP000008711 UP000037069 UP000002282 UP000007801 UP000008792 UP000001292 UP000000803 UP000009192 UP000192221 UP000001819 UP000268350 UP000007798 UP000215335 UP000092553 UP000001070 UP000005204 UP000078541 UP000005203 UP000218220 UP000092461 UP000076407 UP000075900 UP000075886 UP000242457 UP000015104 UP000000305 UP000076858
Pfam
PF00135 COesterase
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
A0A437BB32
A0A2W1BDC8
A0A194RMK8
A0A194Q2A5
A0A212EHX5
A0A1L8D6N7
+ More
A0A139WI26 A0A1I8PGZ6 E0VFS8 A0A1I8MUT4 A0A1B0AJ85 A0A1A9X8I2 A0A1B0B268 Q7PGX1 A0A1A9X427 B4GL70 A0A068F5U0 B4QS57 B3NZ06 A0A0L0BSZ4 B4IUR7 A0A0P8XXC1 B4LXI9 A0A0A9ZGF2 B4IIE9 Q9VDP5 B4K9C7 B6IDZ4 A0A2H1VKL6 A0A0Q9X167 A0A1W4VXD5 W8BSI7 A0A0Q9X8Z0 B5DVH8 A0A3B0KJP9 B4NGA7 A0A232FEM3 A0A0M5J2B0 B4JTH3 H9IY47 A0A437BB17 A0A151JW79 A0A2S2PN03 A0A2S2QDT7 A0A087ZVK3 B9VMQ6 E0VI51 A0A2H1V8H5 A0A0U2LHT4 A0A2A4K584 A0A2W1BEL2 A0A1B0CSA5 A0A1B6CJ17 A0A1B6MDA4 B4M4J0 A0A2C9H8F4 A0A1S4G7C2 A0A1Y9HEJ0 A0A212FJJ8 A0A023F4C2 A0A182Q9A1 A0A1Q3FUD5 A0A194QTD3 A0A2A3E952 A0A2W1BB78 A0A1L8D6I9 A0A087ZUT3 B9VMQ5 A0A2A4JUM4 A0A0M5IZD8 T1L1F5 A0A194Q1D9 A0A1B6KRZ7 A0A2S2R616 E9HH57 E9HH56 Q5TT55 A0A0N8EFR4 A0A1B6D837 A0A164RAU1 A0A0P5XSR9 A0A0P5IJN0 A0A0P6GTR2 A0A0P5MA48 A0A0N8EA47 A0A0P6FXX8 A0A0P5M6G2 A0A0P6FFA3 A0A0P5NL51 A0A0P5QUF6 A0A0P6FQ59 A0A0P6G3F7 A0A0P5RQX5 A0A0P6DGG2 A0A0P6G820 A0A0P6FJW7 A0A0P5WPQ1
A0A139WI26 A0A1I8PGZ6 E0VFS8 A0A1I8MUT4 A0A1B0AJ85 A0A1A9X8I2 A0A1B0B268 Q7PGX1 A0A1A9X427 B4GL70 A0A068F5U0 B4QS57 B3NZ06 A0A0L0BSZ4 B4IUR7 A0A0P8XXC1 B4LXI9 A0A0A9ZGF2 B4IIE9 Q9VDP5 B4K9C7 B6IDZ4 A0A2H1VKL6 A0A0Q9X167 A0A1W4VXD5 W8BSI7 A0A0Q9X8Z0 B5DVH8 A0A3B0KJP9 B4NGA7 A0A232FEM3 A0A0M5J2B0 B4JTH3 H9IY47 A0A437BB17 A0A151JW79 A0A2S2PN03 A0A2S2QDT7 A0A087ZVK3 B9VMQ6 E0VI51 A0A2H1V8H5 A0A0U2LHT4 A0A2A4K584 A0A2W1BEL2 A0A1B0CSA5 A0A1B6CJ17 A0A1B6MDA4 B4M4J0 A0A2C9H8F4 A0A1S4G7C2 A0A1Y9HEJ0 A0A212FJJ8 A0A023F4C2 A0A182Q9A1 A0A1Q3FUD5 A0A194QTD3 A0A2A3E952 A0A2W1BB78 A0A1L8D6I9 A0A087ZUT3 B9VMQ5 A0A2A4JUM4 A0A0M5IZD8 T1L1F5 A0A194Q1D9 A0A1B6KRZ7 A0A2S2R616 E9HH57 E9HH56 Q5TT55 A0A0N8EFR4 A0A1B6D837 A0A164RAU1 A0A0P5XSR9 A0A0P5IJN0 A0A0P6GTR2 A0A0P5MA48 A0A0N8EA47 A0A0P6FXX8 A0A0P5M6G2 A0A0P6FFA3 A0A0P5NL51 A0A0P5QUF6 A0A0P6FQ59 A0A0P6G3F7 A0A0P5RQX5 A0A0P6DGG2 A0A0P6G820 A0A0P6FJW7 A0A0P5WPQ1
PDB
3BE8
E-value=2.05957e-34,
Score=360
Ontologies
GO
Topology
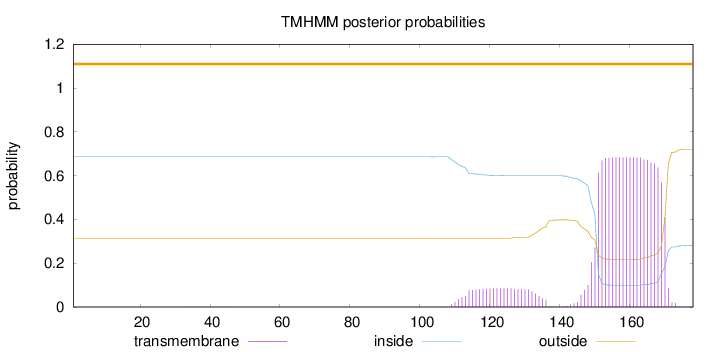
Length:
178
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
15.87626
Exp number, first 60 AAs:
0.00331
Total prob of N-in:
0.68605
outside
1 - 178
Population Genetic Test Statistics
Pi
197.28607
Theta
177.028023
Tajima's D
0.577163
CLR
148.910693
CSRT
0.539623018849058
Interpretation
Uncertain
